 Science Highlights
Science Highlights
CHAPERONE FRAGMENTS AS THERAPEUTIC AGENTS FOR HUNTINGTON DISEASE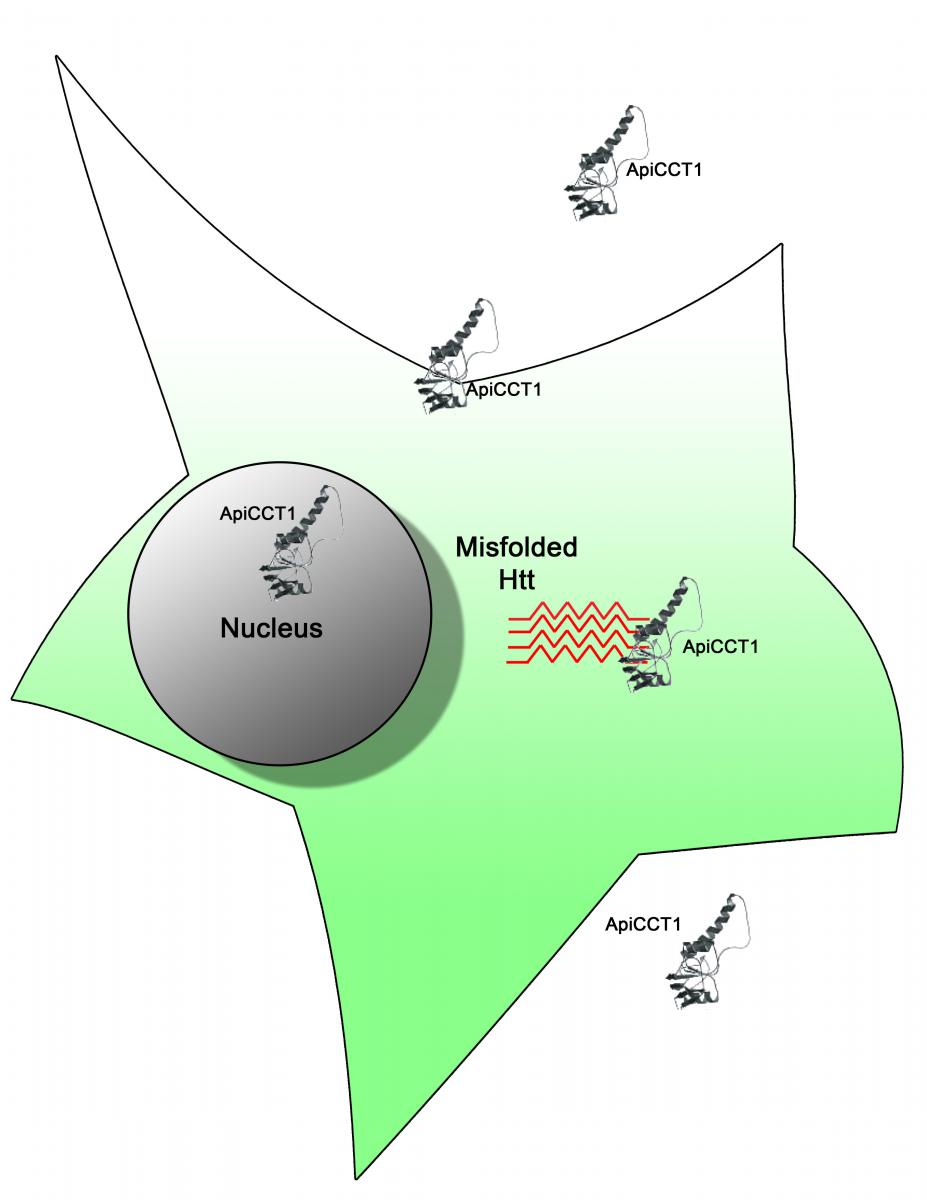
Scientists working on Huntington disease (HD) have uncovered a potential therapeutic strategy to target the abnormal protein aggregates, mHtt, that cause neurodegeneration. Mutations in the HTT gene cause Huntington disease. Aberrant Htt polypeptides misfold and aggregate to form inclusion bodies within neuronal cells. These inclusions are the earliest pathological changes seen in HD affected brains.
Molecular chaperones that assist in proper protein folding are one of the most potent suppressors of neurodegeneration in animal models of human disease, and potential targets to ameliorate inclusion body formation. The TriC chaperonin is a large, 1 mega-Dalton, multi-subunit molecular chaperone that binds newly translated protein fragments and is known to suppress protein aggregation. Focusing their efforts on ApiCCT1, a 20 kilo-Dalton fragment of TRiC, a team led by Drs. Judith Frydman, Steven Potkin, and Leslie Thompson report in their PNAS article, that exogenously delivered ApiCCT1 reduces mHtt aggregation, delays inclusion body formation, and reduces mHtt- mediated neurotoxic effects by increasing cell survival. The team established that ApiCCT1 can penetrate neuronal cell membranes and localize to the nucleus, thus simplifying and eliminating the arduous procedure of expressing ApiCCT1 within the cell.
The investigators, members of the Nanomedicine Development Center for Protein Folding Machinery are supported by the NIH Common Fund Nanomedicine Program.
Reference:
Exogenous delivery of chaperonin subunit fragment ApiCCT1 modulates mutant Huntingtin cellular phenotypes. Proc Natl Acad Sci U S A. 2013 Feb 19; 110(8):3077-82
TUNABLE QUANTUM DOTS FOR MEDICAL IMAGING

Quantum dots are nanocrystals that are activated by and emit specific wavelengths of light. They have been studied for use in many applications; for example, in transistors, solar cells, and LEDs. In recent years, quantum dots (QDs) have been emerging as an excellent way to fluorescently label biological samples, which makes them useful for applications in biomedical imaging and diagnostics. Various fluorescent tags have been used to label biological samples for decades, but QDs are superior because they are brighter and more resistant to degradation by the activating light source. In addition, QDs can be tuned to emit light across a broad range of wavelengths by adjusting their size, shape, and/or energy level. However, one problem that QDs have is that they can be quite large relative to traditional labels, which limits their usefulness in imaging studies. Furthermore, in a given preparation of QDs, there can be variability in their size, stability under illumination, and emission spectra.
Drs. Andrew Smith and Shuming Nie, researchers in the NIH Nanomedicine Center for Nucleoprotein Machines, supported by the NIH Common Fund, developed a new approach to synthesize QDs that permitted unprecedented control over their preparation. This new strategy allowed the researchers to produce very bright and compact QDs that emit wavelengths of light between 700 and 1150 nanometers (nm). Quantum dots with emission spectra in the near to infrared range (700-1700 nm) have advantages, such as lower background interference, better tissue penetration, and enhanced stability. To make them useful in biomedical applications, the QDs were capped with a multilayer shell that enhanced their solubility in water and their stability.
The improved QDs are expected to be a highly effective tool in imaging live cells, as well as other diagnostic applications, and therefore have the potential to be useful in numerous biological applications well beyond the scope of the center. Future work will evaluate their chemical stability and cellular toxicity compared to traditional QDs.
Read more about the Nanomedicine Center for Nucleoprotein Machines
Read more about the Nanomedicine program
Reference:
Smith, A.M. and Nie, S. Bright and Compact Alloyed Quantum Dots with Broadly Tunable Near-Infrared Absorption and Fluorescence Spectra through Mercury Cation Exchange. J. Amer. Chem. Soc. 2011. 133: 24-26. PMID: 21142154.
LIGHT-CONTROLLED PAIN RELIEVER WILL ILLUMINATE MYSTERIES OF ACUTE AND CHRONIC PAIN

Common Fund supported researchers have developed a molecule that can silence pain-sensing neurons using light, opening up new avenues in pain research and the design of novel analgesics. Drs. Dirk Trauner and Richard Kramer, part of the Nanomedicine Development Center for Optical Control of Biological Function, along with their colleagues, have published a study in the February 19, 2012 advance online edition of Nature Methods that describes how a molecule called QAQ can selectively inhibit pain-sensing neurons, or nociceptors. QAQ is a “photoswitch,” a molecule that changes properties when exposed to different wavelengths of light. At one wavelength, QAQ forms an elongated structure that is capable of blocking channels in the membranes of nociceptors. Blocking these channels prevents nociceptors from sending signals, thereby halting the sensation of pain. At a different wavelength, QAQ forms a bent structure that does not block the membrane channels, allowing the nociceptors to function as normal. QAQ affects only nociceptors that are actively transmitting pain signals, unlike local anesthetics that block the functioning of all neurons. This exquisite selectivity, along with precise ability to rapidly turn nociceptors on or off with different wavelengths of light, will be a valuable research tool to study the biology of both acute and chronic pain. It is possible that someday QAQ, or its derivatives, may also be useful as a clinical tool to relieve pain in some patients if a suitable light source can be applied to the affected area. Although much research remains to be done before QAQ can be used as a pain reliever, this study reveals exciting new possibilities for management of pain disorders.
Read the university news release…
Read more about the Nanomedicine Development Center for Optical Control of Biological Function…
Read more about the Nanomedicine program…
Reference:
Mourot A, Fehrentz T, Le Feuvre Y, Smith CF, Herold C, Dalkara D, Nagy F, Trauner D, Kramer RH. Rapid optical control of nociception with an ion-channel photoswitch. Nature Methods, 2012 Feb 19. doi: 10.1038/nmeth.1897 [Epub ahead of print]. PMID: 22343342.
 GETTING PROTEINS INTO THE RIGHT SHAPE
GETTING PROTEINS INTO THE RIGHT SHAPE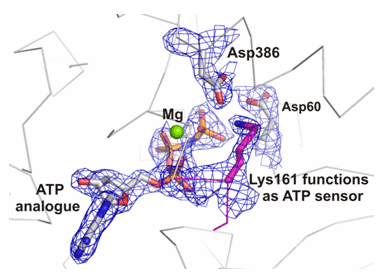
Proteins perform many essential functions that keep cells alive and healthy. But for a protein to carry out its designated function, it must have a specific shape. How do proteins acquire their shapes in the first place? Scientists from the labs of Drs. Jonathan King, Judith Frydman and Paul Adams, part of the Nanomedicine Program’s Center for Protein Folding Machinery, have uncovered new clues about how a type of protein complex called a chaperonin helps fold other proteins into their correct shapes. Published December 23, 2011 in The EMBO Journal, this research may lead to new ways to treat diseases linked to the incorrect folding of proteins, such as Alzheimer’s and Parkinson’s disease.
Chaperonins form a cup shaped structure with a lid. Unfolded proteins enter a protected central cavity and get folded after the lid closes. The question is: how do chaperonins do their job? To answer this question, the researchers used extremely powerful X-ray beams produced by the Advanced Light Source at the Lawrence Berkeley National Laboratory to determine the structure of a critical control region within the chaperonin. This region, called the nucleotide sensing loop (NSL), detects the binding of a molecule called ATP to a specific part of the chaperonin. The NSL then communicates information about the ATP binding to the rest of the chaperonin complex as a signal to all the chaperonin components to rotate together in a coordinated fashion. When this happens, the protein inside the cavity gets folded into the correct shape.
The high-resolution structure revealed the exact chemical interactions between the ATP molecule and the NSL. Further experiments demonstrated that the NSL also regulates the chemical reaction in which the ATP is broken down, released, and new ATP attaches to the chaperonin to start a new round of protein folding. The knowledge of how this process takes place may enable researchers to engineer chaperonins with modifications to the NSL that alter the protein folding activity, possibly allowing the engineered chaperonin to reduce the cellular accumulation of incorrectly folded proteins in disease.
Read the news release….
Read more about the Center for Protein Folding Machinery….
Read more about the Nanomedicine program…
Reference:
Pereira JH, Ralston CY, Douglas NR, Kumar R, Lopez T, McAndrew RP, Knee KM, King JA, Frydman J, and Adams PD. Mechanism of nucleotide sensing in group II chaperonins. The EMBO Journal, Dec 23, 2011. 31(3): 731-740. PMID: 2219370.
 NANOSCALE IMAGING OF PROTEIN AGGREGATES IN HUNTINGTON'S DISEASE
NANOSCALE IMAGING OF PROTEIN AGGREGATES IN HUNTINGTON'S DISEASE
Protein aggregation is a hallmark of neurodegenerative diseases such as Huntington’s, Alzheimer’s and Parkinson’s disease. The proteins responsible for neurodegeneration misfold and form clumps, or aggregates. In the case of Huntington’s disease, Htt aggregates later change shape to form branches and give rise to other fibrils by more clumping or breakage of the initial fibrils. Although disease severity has been related to the size of these aggregates, it is unclear if aggregates play a causative role or a protective role in disease formation. It is thought that morphological changes of aggregates might explain the disease mechanism; if so, studying aggregate formation will aid development of potential therapies for diseases such as Huntington’s. Using super-resolution (SR) fluorescence imaging, scientists at Stanford University distinguished single molecules of Huntingtin (Htt) protein with nanometer precision within protein aggregates.
SR imaging has emerged as a powerful new tool to visualize fine biomolecular architecture at the nanoscale level. Unlike the long fibrils seen in other neurodegenerative diseases, Huntington’s Htt aggregates form short, intricate branches, challenging the limits of optical imaging techniques, due to their fine structure. The investigators attached Htt aggregates to a photoactivatable fluorescent dye that emits light in a fluctuating manner, giving a “blinking” appearance. This allowed localization of single molecules that were twelve nanometers apart within the aggregates. In this work, Moerner and colleagues have successfully applied the power of this high resolution imaging technique to a germane biological problem, thereby providing the first single molecule snapshots of the aggregates associated with Huntigton’s disease. Earlier single molecule studies had been unable to hone in at the nanometer dimension. With this innovative application of SR imaging to observe Htt aggregates, scientists can next move on to image protein aggregates in real-time to understand Htt and other neurodegenerative proteins in their cellular environments.
The work was supported by the NIH Nanomedicine Development Center for Protein Folding Machinery, which is developing nanoscale approaches to correct misfolded proteins in neurodegenerative diseases.
Publication citation and link:
Duim WC, Chen B, Frydman J, Moerner WE (2011). Sub-Diffraction Imaging of Huntingtin Protein Aggregates by Fluorescence Blink-Microscopy and Atomic Force Microscopy. Chemphyschem.12:1-4.
 NANOPARTICLES DELIVER COMBINATION CARGOS DIRECTLY TO CANCER TARGETS
NANOPARTICLES DELIVER COMBINATION CARGOS DIRECTLY TO CANCER TARGETS
Successful treatment for cancer requires efficient delivery of potent drugs to kill tumor cells with minimal side effects. In a study* published online in the journal Nature Materials in April, investigators constructed synthetic “protocells” that were used to kill liver tumor cells without adversely affecting healthy cells. Protocells are made by enclosing highly porous silica nanoparticles, 100-150 nanometers in diameter, with a lipid bilayer thus mimicking a cell membrane. The bilayer includes protein ligands that specifically target tumor cells, and the porous core is preloaded with anti-cancer drugs. Upon binding to its target, the protocell enters the tumor cell and releases its cargo, thereby killing the cancerous cell. Protocells can carry high concentrations and different combinations of cargo, such as drugs, small interfering RNAs, and other toxins. The cargo capacity and time course of release can be controlled by changing the pore size and chemistry of the silica core, but the protocell is designed to release the cargo only upon entry into the target cell. In the future, protocells may be designed to efficiently and effectively target and kill various types of cancer cells, depending on the cargo loaded inside and the targeting molecules used on the surface of the protocell. However, before using in human studies, protocells will first need to be tested in animal models.
The investigators of this study were supported in part by the National Center for Design of Biomimetic Nanoconductors, which is one of eight centers of the NIH Nanomedicine Program, funded by the NIH Common Fund.
Reference:
*Ashley CE, Carnes EC, Phillips GK, Padilla D, Durfee PN, Brown PA, Hanna TN, Liu J, Phillips B, Carter MB, Carroll NJ, Jiang X, Dunphy DR, Willman CL, Petsev DN, Evans DG, Parikh AN, Chackerian B, Wharton W, Peabody DS, Brinker CJ (2011). The targeted delivery of multicomponent cargos to cancer cells by nanoporous particle-supported lipid bilayers. Nature Materials 10: 389-397.
Read more about the Nanomedicine program…
Up to Top
 NEURONS FILTER OUT IRRELEVANT INFORMATION
NEURONS FILTER OUT IRRELEVANT INFORMATION
 Scientists have discovered how a small subset of neurons in the zebrafish brain has a big impact on an important behavior—the ability to hunt down prey. In a study published in the October 29th issue of Science, Dr. Herwig Baier of UC San Francisco and Dr. Ehud Isacoff of UC Berkeley, investigators in the Common Fund’s Nanomedicine program, use a novel fluorescent reporter of nerve cell activity to uncover how zebrafish can spot a tiny, one-celled paramecium against a complex visual background. The optic tectum in zebrafish is a layered structure that receives signals from the eye in the superficial layer, and sends signals from the deeper layer out to motor areas of the brain that control movement. Drs. Baier and Isacoff and colleagues demonstrate that a group of neurons in the optic tectum called superficial inhibitory neurons (SINs) acts to "filter out" large background patterns, allowing the tectum to send specific messages to motor areas about small, moving objects like prey. Silencing or destroying SINs eliminates this filtering, and impairs the zebrafish’s ability to catch prey. The selective filtering of irrelevant background information is found throughout the brain of many animals, including humans, but relatively little is known about the individual nerve cells that underlie this process. This study offers important insight about how circuits of nerve cells can provide background filtering, and suggests similar mechanisms may be found in human brains.
Scientists have discovered how a small subset of neurons in the zebrafish brain has a big impact on an important behavior—the ability to hunt down prey. In a study published in the October 29th issue of Science, Dr. Herwig Baier of UC San Francisco and Dr. Ehud Isacoff of UC Berkeley, investigators in the Common Fund’s Nanomedicine program, use a novel fluorescent reporter of nerve cell activity to uncover how zebrafish can spot a tiny, one-celled paramecium against a complex visual background. The optic tectum in zebrafish is a layered structure that receives signals from the eye in the superficial layer, and sends signals from the deeper layer out to motor areas of the brain that control movement. Drs. Baier and Isacoff and colleagues demonstrate that a group of neurons in the optic tectum called superficial inhibitory neurons (SINs) acts to "filter out" large background patterns, allowing the tectum to send specific messages to motor areas about small, moving objects like prey. Silencing or destroying SINs eliminates this filtering, and impairs the zebrafish’s ability to catch prey. The selective filtering of irrelevant background information is found throughout the brain of many animals, including humans, but relatively little is known about the individual nerve cells that underlie this process. This study offers important insight about how circuits of nerve cells can provide background filtering, and suggests similar mechanisms may be found in human brains.
Reference:
Del Bene F, Wyart C, Robles E, Tran A, Looger L, Scott EK, Isacoff EY, Baier H. Filtering of visual information in the tectum by an identified neural circuit. Science, 2010 Oct 29; 330 (6004): 669-73. PMID: 21030657.
Read more about the Nanomedicine program…



