2007 Awardees

Kjersti M. Aagaard-Tillery
Baylor College of Medicine
Project Title: Characterization of the Fetal Primate Epigenome and Metabolome Under In Utero Conditions of Maternal Obesity
Grant ID: DP2-OD001500
Obesity causes substantial social, economic and health burdens. The rate of obesity is escalating disproportionately in children (infants to young adults). This rapid increase is unlikely to be due to environment or genetics alone. Accumulating evidence from our laboratory and others suggests that adult metabolic diseases originate in utero, and likely occur through the reprogramming of gene expression via epigenetic changes in chromatin structure (an altered "histone code"). Of interest, we have observed in a rodent transgenerational model of intrauterine growth restriction (IUGR) that a diet supplemented with essential nutrients, yet unaltered in its caloric content, prevents adult metabolic disease and is associated with abrogation of reprogrammed gene expression. However, although such established models in rodents demonstrate that fetal alterations in the histone code are involved in the persistence and conveyance of the altered postnatal phenotype, little is known about the effects of maternal diet and resultant obesity on primate fetal biology. We hypothesized that a high fat diet in non-human primates would induce changes in hepatic chromatin structure resulting in altered expression of fetal genes critical to the development of childhood and adult obesity. Based on our preliminary data, the focus of this proposal is to apply developed high throughput technology (comparative epigenomics and metabolomics) to decipher the primate epigenome and metabolome in the obese maternal environment and then measure the impact of supplementation on the differentially altered epigenome and resultant disease. The novel innovation and significance resides within its potential to provide (1) an expanded understanding of the mechanism through which a maternal high fat diet reprograms primate gene expression and (2) a simple intervention (essential nutrient supplementation with neither diet nor behavioral modification) with tremendous potential impact given the current obesity epidemic and the lack of efficacious therapeutics.

Ryan C. Bailey
University of Illinois Urbana-Champaign
Project Title: Personalized Clinical Diagnostics and Beyond: Integrated Ring Resonator Arrays
Grant ID: DP2-OD002190
Paradigm shifts in biology are often catalyzed by innovations in measurement technologies. Genomics and proteomics have revolutionized biology but would not have been possible without developments in capillary sequencing, cDNA microarrays, and mass spectrometry, amongst other enabling technologies. Cancer biology has significantly benefited from the molecular-level detail provided by these tools, allowing elucidation of many perturbations underlying disease onset and progression. Unfortunately, many of the same measurement approaches are not applicable in the clinical setting and thus physicians do not have access to the same detailed biochemical information enjoyed by the academician. As a result, despite our increased knowledge of the molecular bases of cancer, the translation to clinical medicine has lagged significantly behind. This proposal describes a revolutionary biological analysis technology which has the potential to profoundly change the face of clinical medicine and beyond. High density arrays of extraordinarily sensitive integrated microring resonators will allow many gene and protein signatures to be simultaneously quantitated from a single patient sample. Distinguishing features of this technology include: sensitivity allowing PCR-less gene and single protein detection, label-free and real time operation, ultra-high scalability (>50,000 sensors/cm2), automated microfluidic operation, and commercially validated manufacturability via CMOS-compatible processing. To demonstrate the power of this technology, we will generate a molecular disease fingerprint allowing differentiation between three clinically indistinguishable yet biochemically distinct disease pathways underlying the deadly brain cancer glioblastoma multiforme. Importantly, each of these pathways is known to respond effectively to different therapeutic agents, thus personalized diagnosis equates to personalized treatment. We will also utilize this enabling technology to provide insight into profound questions surrounding post-transcriptional gene regulation and heterogeneity within the secreted responses of individual immune cells. This technology promises to broadly impact the landscape of the biomedical sciences, both meeting the clinical diagnostic challenges of today and pioneering the paradigm-shifting discoveries of tomorrow.

Edward S. Boyden
Massachusetts Institute of Technology
Project Title: Novel Tools and Principles for Precisely Controlling Brain Activity
Grant ID: DP2-OD002002
The finding that many neurological and psychiatric disorders are associated with abnormal neural activity in specific brain circuits raises the optimism that a precise, flexible technology for controlling neural activity would enable the systematic treatment of many brain disorders. I here propose to take a top-down approach to developing a new tool for noninvasive, focal, deep-targetable control of brain circuits. I also propose to discover informed principles governing the use of these tools to control activity in a diversity of neural circuits relevant for different illnesses. Through far-reaching collaborations, I will not only invent these tools and discover how to use them, but lead their translation into clinical application. I believe that this is the right time and place to tackle this intellectual challenge, given my unique training, as well as my proven abilities to synthesize new insights from disparate fields, and to lead interdisciplinary teams into new territory.

Frances A. Champagne
Columbia University, New York Morningside
Project Title: Epigenetic Mechanisms Mediating the Inheritance of Reproductive Behavior
Grant ID: DP2-OD001674
Natural variations in mother-infant interaction have profound effects on offspring development and behavior. In particular, there is evidence for the influence of postpartum care on maternal behavior itself, such that low levels of maternal care are exhibited by females who received less care in infancy. This transmission of maternal behavior from mother to daughter has been previously associated with epigenetic regulation of estrogen receptor (ER) a in hypothalamic brain regions. The purpose of the proposed project is to explore these epigenetic effects in the context of reproductive behavior and develop convergent sources of evidence for the transgenerational influence of maternal care. To achieve this goal, the maternal regulation of social, sexual and maternal behavior will be explored in a rodent model. This will provide an experimental design for determining potential shifts in reproductive strategy in response to early environmental experiences. The reversibility of these maternal effects will be explored through pharmacological manipulation of DNA methylation within the promoter region of the ERa gene. The role of DNA methylation in the generational transmission of environmental experiences that occur beyond the postpartum period will also be investigated in response to gestational stress and juvenile exposure to differential levels of social interaction. Finally, ecologically valid paradigms will be developed in which transgenerational effects on reproductive behavior can be determined in a mouse model. These paradigms will involve 1) comparison of communally vs. non-communally reared offspring and grand-offspring, 2) caloric food restriction during lactation and 3) comparison of offspring born to either inexperienced or experienced mothers. Overall, this project will provide an innovative approach to both the study of inheritance and the role of epigenetic mechanisms in sustaining environmental effects within and across generations. Moreover, it will provide an experimental design for elucidating the occurrence of reproductive trade-offs in mammals.

Sean S. Davies
Vanderbilt University
Project Title: Transformed Probiotic Bacteria for Treatment of Chronic Diseases
Grant ID: DP2-OD003137
The continual increase in the number of patients suffering from chronic medical conditions that require long term treatment with therapeutic drugs such as hypercholesterolemia, hypertension, diabetes, and obesity has proven a tremendous economic burden, both to individuals and to health care systems. The traditional approach to drug production has been chemical synthesis of the needed compounds, then purification, formulation, and distribution. We propose to investigate a novel approach to long-term drug production and delivery: using probiotic intestinal bacteria transformed to express the required drug and inoculated into a patient for chronic colonization and therapeutic production. This approach essentially takes the notion of gene therapy and rather than altering the genomic DNA of the patient, instead alters the DNA of the patient's commensal bacteria, a far more tractable system. Probiotic intestinal bacteria such as members of the Lactobacillus family are routinely added to foods such as dairy products, have essentially no pathology, and can be readily transformed with exogenous DNA. Lactobacillus transformed with therapeutic proteins and peptides or sets of enzymes to synthesize specific drugs may therefore represent a versatile platform for sustainable therapy. We will determine the optimal strains of Lactobacillus for expression in mice as a model organism and perform additional engineering of the strain as necessary to ensure persistence in the intestinal tract and regulated expression of peptide or proteins. We will also develop methods for rapidly depleting the transformed bacteria without damaging other commensal bacteria, as a safety precaution against the advent of any unexpected adverse responses during the use of these therapeutically transformed bacteria. We will then examine the effect on atherosclerosis-prone mice of expressing peptides with known therapeutic actions. Our final aim will be to test the feasibility of producing small molecule drugs such as lovastatin in vivo by transforming the bacteria with the required synthetic enzymes.

Pedro Fernandez-Funez
University of Florida
Project Title: Mechanisms of Prion Misfolding
Grant ID: DP2-OD002721
Prion diseases are a group of aggressive, lethal and incurable neurodegenerative disorders. The hallmark pathological event in these maladies is the misfolding, aggregation and brain deposition of “scrapie” Prion protein (PrPSc), which leads to spongiform degeneration of brain neurons. Unfortunately, a major gap exists in our understanding of how the conformational conversion of PrP ultimately kills neurons. Recent in vitro studies suggest that molecular chaperones may be key factors mediating PrP conversion and neuronal dysfunction. However, the functional relevance of this finding is unknown. My central hypothesis is that targeted expression of molecular chaperones can suppress PrP misfolding and PrP-mediated neurodegeneration. To that end, I created a novel and powerful Drosophila model of sporadic prion disease in which wild type PrP from Hamster converts into PrPSc–like conformations and causes spongiform degeneration. Supporting my hypothesis, human Hsp70 prevents PrP misfolding and protects against PrP-dependent neurotoxicity in transgenic flies. The overall goal of this project is to define the role of molecular chaperones in PrP misfolding by a multidisciplinary approach that combines the power of Drosophila genetics with mammalian cellular systems and mice. My specific aims are: 1) Genetic, biochemical and pharmacological approaches to study Hsp70 protection against PrP misfolding and neurotoxicity in Drosophila; 2) Determination of the ability of the Unfolded Protein Response components XBP1s and Grp58 to suppress PrP misfolding and neurotoxicity in Drosophila; 3) Role of molecular chaperones in prion replication in simplified mammalian systems, and 4) Potential therapeutic role of Hsp70 in mouse models of prion diseases. I anticipate that our studies will contribute to better understand the molecular basis underlying PrP conversion. In addition, by exploring the role of chaperone-inducing compounds in flies and mice, I may discover effective and innovative therapeutic interventions for treating these devastating and yet incurable diseases.

Sarah M. Fortune
Harvard University (School of Public Health)
Project Title: Variation in M. tuberculosis in Response to Host Selection
Grant ID: DP2-OD001378
All pathogens that cause chronic infections must avoid clearance by the host immune response. Many have complex mechanisms to rapidly generate diversity in critical antigens. Mycobacterium tuberculosis chronically infects one third of the worlds' population and similarly must avoid clearance by the host immune system. However, there is currently little understanding of whether M. tuberculosis, like so many other pathogens, diversifies in vivo to escape host immune selection. In this proposal, we will test the hypothesis that M. tuberculosis varies, either genetically or epigenetically, during the course of infection and that this variation contributes to the ability of the bacteria to avoid clearance by the host immune response. We will use new genomics technologies—low cost genome sequencing and expression profiling—to systematically assess genetic and epigenetic variation in bacteria selected in a simple experimental model of disease chosen to create different immune pressures on the bacteria —mice of different MHC haplotypes. In these studies, we expect to provide fundamental insights into the mechanisms and targets of diversifying immune selection in M. tuberculosis.

Levi Alexander Garraway
Dana-Farber Cancer Institute
Project Title: Defining Melanoma Therapeutic Avenues by Integrative Functional Genomics
Grant ID: DP2-OD002750
Although tumor classification and patient stratification based on genomic criteria offers tremendous clinical potential, discerning critical effectors of tumor genetic alterations—and developing robust therapeutic avenues to intercept them—represents a formidable obstacle to translational oncology. Recent advances in large-scale ‘perturbagen' approaches (e.g., viral RNAi and small molecule screening) hold great promise to alleviate such bottlenecks; however, their systematic application in cancer biology would benefit markedly from robust in vitro cancer models that fully encompass the genomic diversity manifest in patients. Malignant melanoma offers a rich avenue in this regard: unlike other solid tumors, cells from this lethal malignancy are readily cultured in vitro, thereby providing a diverse and tractable system for genomic and functional studies. Accordingly, we propose to apply pooled RNAi and small molecule microarray screening sequentially to a panel of genetically characterized and patient-derived melanoma cell lines. We will assemble a lentiviral library targeting all expressed genes located within the portion of the genome that is amplified in melanoma, and perform pooled RNAi screening across a panel of 20 melanoma lines representative of the most prevalent melanoma genomic alterations. ‘Hits' (e.g., selectively depleted shRNAs) will be correlated with genomic patterns to identify (onco)gene targets of melanoma amplifications. We will validate the most promising target genes using a series of cell survival assays in vitro (in arrayed RNAi format) and tumor formation assays in vivo (using shRNA ‘mini-pools'). Finally, we will identify candidate ligands that bind the top target (onco)protein candidates by performing small-molecule microarray screens using epitope-tagged protein constructs. If successful, this project should elaborate a spectrum of target proteins and potential lead compounds linked to common genetic changes in melanoma. Moreover, these efforts should inform a “platformizable” integrated approach applicable to all cancers for which tractable in vitro models exist.

Tawanda Gumbo
University of Texas Southwestern Medical Center
Project Title: Efflux Pump Inhibitors to Reduce Duration of Antituberculosis Therapy
Grant ID: DP2-OD001886
Tuberculosis has devastated mankind for millennia. Current therapy is based on a belief that Mycobacterium tuberculosis in lung cavities exists as one of three populations: rapidly growing bacilli in areas of high oxygen that are most effectively killed by isoniazid, slowly growing bacilli under acidic conditions that are killed by pyrazinamide, and non-replicating bacilli under low oxygen tension that are killed most effectively by rifampin. We and others have recently demonstrated that parts of this belief may be incorrect. In the current proposal, I provide evidence of a central role for drug-efflux pumps to each of the first line anti-tuberculosis drugs. These drug-efflux pumps may lead to high level resistance. However, even low-level resistance efflux pumps may still provide a crucial survival advantage that enables bacilli to survive antibiotic exposure. I propose that the differential induction of drug-efflux pumps under different microenvironmental conditions may be responsible for selective effect of first line antituberculosis compounds under different oxygen and pH conditions. Induction of these pumps leads to increased mutation rates and emergence of resistance in vitro and in vivo. Inhibition of the efflux pumps by a common inhibitor will lead to acceleration of M. tuberculosis microbial kill, whether the bacilli are replicating or not. Three inexpensive efflux pump inhibitors, which are currently commercially available off patent, will be utilized to test these hypotheses in vitro and in vivo. After that, preclinical pharmacokinetic-pharmacodynamic studies will be performed. The drug concentrations of the efflux pump inhibitors best able to shorten duration of standard antituberculosis therapy will then be identified. Using population pharmacokinetics and pharmacogenomics, these results will be translated via Monte-Carlo simulations to identify (a) optimal dose of inhibitor best able to achieve this in humans and (b) the optimal duration of this therapy in humans. These results will then be prospectively validated.

Nir Hacohen
Massachusetts General Hospital
Project Title: Revealing Pathogen-Sensing Pathways Using RNAi Libraries
Grant ID: DP2-OD002230
The long-term goal of the proposed work is to apply a genome-wide lentiviral RNAi library that we have developed to dissect innate immune pathways in mammals. In this proposal, we describe a novel experimental strategy to explore an elaborate and ancient sensory system that detects pathogen-derived nucleic acids. The nucleic-acid sensory system has been implicated in the detection of most pathogens and in the initiation of two major autoimmune diseases, systemic lupus erythematosus and rheumatoid arthritis. Our two objectives are to identify a set of unknown DNA sensors and their pathways, and to understand how DNA and RNA sensors avoid being inappropriately activated by host DNA and RNA. To identify genes and proteins in the unknown DNA-sensing pathways, we will: (1) perform a rapid unbiased genome-wide pooled RNAi screen to identify the strongest hits; (2) apply a more sensitive arrayed RNAi screen to test the role of a pre-selected subgenome library of genes; (3) purify dsDNA-binding proteins and use mass spectrometry to identify these proteins; (4) characterize the functions of newly identified genes required for DNA-sensing. To explore the mechanisms that inhibit the sensing of self nucleic acids, we will: (1) perform a pooled RNAi screen to identify blockers of spontaneous activation in response to endogenous nucleic acids; (2) perform an arrayed screen to identify genes involved in detection of dying cell nucleic acids by dendritic cells. By understanding this detection system in depth, we will gain insight into the perpetual struggle between parasitic elements and their hosts, and the risks of autoimmunity in response to the host's own nucleic acids. In addition, the results of the studies will help in the rational development of vaccines using adjuvants that target nucleic acid sensors.

Ekaterina Heldwein
Tufts University Boston
Project Title: Structural and Mechanistic Studies of Herpesvirus Entry into Host Cells
Grant ID: DP2-OD001996
Herpesviruses are human pathogens that infect their hosts for life, causing cold sores, genital herpes, blindness, encephalitis, cancers, and life-threatening conditions in immuno-compromised individuals. The goal of my research is to understand in atomic-level detail how herpesviruses enter host cells. Such information will be invaluable in designing anti-herpesvirus therapeutics to combat both viral infections and cell-cell spread. The herpesvirus cell-entry mechanism is very complex. Whereas other enveloped viruses use a single protein to effect cell entry, all herpesviruses require at least three proteins: gB, gH, and gL. These three proteins are thought to accomplish the fusion of viral and cell membranes – a pivotal step in viral entry – but their exact functions are obscure. I aim to determine how the gB and gH/gL proteins of Herpes Simplex Virus (HSV) work together to accomplish membrane fusion and how the signal from the receptor-binding protein, gD, triggers the membrane-fusion machinery. Uncovering how these proteins work in HSV infection will also reveal their functions in other herpesviruses because the membrane-fusion machinery, i.e., gB, gH, and gL, is highly conserved. My approach will combine the power of x-ray crystallography to determine the structures of individual proteins with the rigor of other biophysical and biochemical techniques to study their interactions. The outcome of this research will be a thoroughly determined and finely detailed picture of the herpesvirus-mediated fusion of the viral and cell membranes. Previous studies of virus-mediated membrane fusion in single-component systems have provided many crucial insights into the general mechanism of membrane fusion, involved in many normal cellular processes. But the complex multiprotein fusion machinery of herpesviruses is a better model for the regulated, multi-component cellular membrane fusion. Therefore, if we establish how herpesvirus glycoproteins interact to drive membrane fusion during cell entry, we will advance fundamentally our mechanistic understanding of membrane fusion.

Konrad Hochedlinger
Massachusetts General Hospital
Project Title: Reprogramming of Somatic Cells by Defined Factors
Grant ID: DP2-OD003266
Nuclear reprogramming defines the dedifferentiation of adult cells into pluripotent embryonic cells and has enormous therapeutic potential as it allows generating genetically matched cells from patients for cell therapy. Reprogramming has so far been achieved by nuclear transfer into oocytes and by cell fusion between embryonic cells and somatic cells, two approaches that have serious technical or ethical limitations. Based on recently published observations, we have generated so-called induced pluripotent stem (iPS) cells directly from fibroblasts by retroviral overexpression of the transcription factors Oct4, Sox2, c-myc and Klf4. In contrast to the previously reported iPS cells, our iPS cells were indistinguishable from ES cells in their epigenetic state and developmental potential. Several crucial questions were raised by these findings; (i) what is the sequence of molecular changes that accompany nuclear reprogramming, (ii) what is the kinetics of reprogramming and does it require cell division, (iii) are different cell types at different differentiation stages equally amenable to reprogramming, and (iv) can human cells be reprogrammed into iPS cells? Resolving these questions will be critical for understanding the molecular nature of nuclear reprogramming and may lead to strategies that allow efficient reprogramming of patient's cells into pluripotent cells. The current limitations to solve these questions are the low efficiency of direct reprogramming and the inability to follow reprogramming in real time. We will tackle these questions by generating “reprogrammable mice” in which every single cell can be reversibly induced to express the four factors at levels necessary for reprogramming, and by attempting to reprogram human cells. The goals of this proposal are thus to determine (i) the robustness and kinetics of reprogramming, (ii) the hierarchy of transcriptional and epigenetic changes that accompany nuclear reprogramming, (iii) the responsiveness of different cell types to the four factors, and (iv) the feasibility of human reprogramming.

Kristen C. Jacobson
University of Chicago
Project Title: From Neighborhoods to Neurons and Beyond
Grant ID: DP2-OD003021
The purpose of this three-phase study is to conduct a multidisciplinary-based investigation of the effects of individual, family, peer, and neighborhood characteristics on individual differences in adolescent problem behavior. Phase I consists of an in-school assessment of approximately 7,000 6th – 8th graders in 10 socioeconomically and racially and ethnically diverse schools in the Chicago Public School system. The purpose of Phase I is to obtain data on environmental and psychosocial factors that may account for socioeconomic and racial and ethnic differences in problem behavior. Phase II consists of a target sample of 400 full-, half- and unrelated sibling pairs drawn from the original Phase I study who, along with their primary caregivers, will be brought to the University of Chicago for detailed neuroscience and behavioral assessments. The primary purpose of Phase II is to identify the environmental, biological, and psychosocial variables that account for individual differences in problem behavior, and to further determine whether these effects are genetically or environmentally mediated. Phase III consists of an fMRI investigation using a target sample of 60 concordant and 40 extreme discordant sibling pairs drawn from the Phase II study. The primary purpose of Phase III is to identify the neurobiological substrates that account for within-pair differences in behavior, and to determine the extent to which nonshared environmental factors account for individual differences in neurobiological functioning. A small pilot study of 20 extremely discordant sibling pairs from Phase III will investigate whether environments alter gene expression via changes in DNA methylation. Because this nested design uses a bioecological framework to measure risk and protective factors at multiple levels of analysis- “from neighborhoods to neurons and beyond”- the resulting data uniquely allows for the investigation of mediating and moderating effects among environmental, psychosocial, biological, and genetic factors on individual differences in problem behavior.

Joanna L. Jankowsky
Baylor College of Medicine
Project Title: Selective Neuronal Silencing to Study Cognitive Decline in Alzheimer's Disease
Grant ID: DP2-OD001734
Our understanding of neurodegenerative diseases is currently hindered by lack of a firm neurobiological link between the patient's symptoms and the underlying neuropathology. To advance, we must identify not only key biochemical changes, but also how these changes alter the function of specific circuits to cause neurological symptoms. I seek to understand how impairment of particular circuits initiates early symptoms of Alzheimer's disease (AD), and how addition of further dysfunction leads to the disease's ultimate decline. I will apply a new method of selective neuronal silencing in transgenic mice to examine the behavioral impact of inactivating neuronal circuits damaged in AD. My postdoctoral laboratory has developed a novel chloride channel that responds specifically to ivermectin by producing hyperpolarization that results in selective, reversible suppression of neuronal activity. I will use my expertise in transgenic technology to create a mouse in which the ivermectin channel is conditionally expressed under control of Cre recombinase. Mating this mouse to animals expressing Cre in selected neuronal populations will allow those cells to be silenced with systemic ivermectin. My goal is to explore the function of adult-born hippocampal neurons, as this population is severely diminished in mouse models for AD. I will examine the role of these cells in learning and memory by selectively silencing them at critical times in the acquisition, consolidation, and recall of new information. Additional studies will address the effect of silencing on the migration, morphology, and survival of these adult-born cells. My long-term plans are to examine the behavioral impact of silencing other circuits damaged later in the course of disease to understand how diminished activity in multiple domains results in the progressive cognitive decline of AD. In the process, I will generate a transgenic mouse for selective neuronal silencing that will be broadly useful to the neuroscience community.

Alan Jasanoff
Massachusetts Institute of Technology
Project Title: Genetically-Controlled MRI Contrast Agents for Functional Brain Imaging
Grant ID: DP2-OD002114
Understanding how the brain controls behavior is one of the outstanding scientific problems of today. The methods most needed to study neural mechanisms of behavior will combine the noninvasiveness and whole-brain coverage of magnetic resonance imaging (MRI) with the precision of cellular recording techniques like electrophysiology and fluorescence microscopy. Here we propose to develop a new set of neuroimaging techniques that approach this ideal. The methods rely on the use of protein-based sensors that report aspects of neural function and may be targeted to specific cells or cell types. Unlike protein sensors developed previously for optical imaging, the sensors we propose to create will incorporate MRI contrast agents, meaning that they can be monitored across entire, intact brains; genetic targeting will allow neural activity of defined “circuit elements” to be identified and integrated into explanatory models of brain function. In the first two Specific Aims, we propose to form and apply genetically-encoded calcium-sensitive contrast agents based on the iron storage protein ferritin (Ft). Our preliminary data and recently published results indicate feasibility of the design, which we will test in rats once it has been validated in cell culture. Ftbased sensors may not offer enough sensitivity for functional imaging of sparse or subtle changes in neuronal physiology, however. To address this, we propose in Aims 3-4 to develop a second genetically controlled system in which markers of neuronal activity are transduced and amplified by enzymes into robust signals that may be read out using powerful exogenous contrast agents. This program is an ambitious and high-impact endeavor that, once accomplished, will change the way researchers study the brain. The PI and his laboratory have the diversity of experience, record of effective collaborations, and risk-taking spirit that will allow them to introduce a new generation of brain measurements and empirical approaches in neuroscience.

Mark D. Johnson
Brigham and Women's Hospital
Project Title: MicroRNA Biogenesis and the Cancer Proteome
Grant ID: DP2-OD002319
Recent studies in human cancers have revealed defects in microRNA biogenesis that promote tumor aggressiveness and decrease survival via mechanisms that are poorly understood. Much of the control of protein expression by microRNAs occurs without alterations in mRNA expression, and single microRNAs may target dozens of mRNAs in a tissue specific manner. Thus, this broad-based decrease in microRNA synthesis presents a challenge to those trying to understand its downstream molecular effects on carcinogenesis. Here we propose a new strategy that will comprehensively identify the mechanisms by which defects in microRNA biogenesis decrease cancer survivorship. This approach involves the integration of genome-scale high throughput quantitative mass spectrometry analysis of the cancer proteome with genome-wide microRNA, mRNA and DNA analyses of human brain tumors (glioblastomas) that have intact or defective microRNA biogenesis. Novel algorithms that incorporate clinical variables will be used to generate an integrated genomescale view of identified differences, and clinically-related targets will be investigated further using in vivo and in vitro models. Preliminary studies in our laboratory using aCGH and mRNA microarrays revealed deletion at the DICER1 locus and decreased DICER1 mRNA expression in a subset of glioblastomas, and this correlated with decreased microRNA expression and survival. Genomewide analysis identified numerous microRNAs that were differentially expressed between glioblastomas with low versus high DICER1 expression. Cdk6, which promotes cell cycle progression, was a predicted target of several of these microRNAs. DICER1 knockdown in glioblastoma cells increased cell growth and upregulated Cdk6 protein expression. Importantly, Cdk6 protein was overexpressed in glioblastomas with low DICER1 expression, suggesting that Cdk6 upregulation may be one mechanism by which defective microRNA biogenesis contributes to increased tumor aggressiveness. This innovative and ambitious project will integrate mass spectrometry proteomics, genomics and clinical variables to comprehensively identify the mechanisms underlying the decreased cancer survivorship associated with dysregulated microRNA biogenesis.

Manuel Llinas
Princeton University
Project Title: Novel Antimalarial Strategies Using Metabolomic Network Discovery
Grant ID: DP2-OD001315
Malaria is a major global health issue affecting over half a billion people and resulting in 3-5 million deaths annually. This disease is caused by parasites of the genus Plasmodium with P. falciparum being the most lethal species. The host-pathogen relationship between Plasmodium and the host red blood cell is responsible for all clinical manifestations of the malaria disease and is a continuous 48-hour cycle that can be faithfully reproduced in the laboratory. Despite over a century of research on malaria, it continues to be a major health problem largely because drug-resistant parasites are on the rise, circumventing long-efficacious drug treatments. Thus, there is a renewed urgency to identify novel chemotherapeutics to treat this disease. This proposal aims to provide the first global analysis of the metabolic host-pathogen interactions for Plasmodium falciparum as a means to identify novel drug targets. The metabolic pathways encoded in any pathogen genome define the repertoire of chemical processes that it can autonomously regulate. All other metabolites must be taken up from the host cell or metabolized from precursors available through the host. Therefore, the host cell and pathogen are intimately linked through the reliance of the pathogen on the host for nutrients. The genome of P. falciparum suggests that this organism is biochemically unique: 60% of its genome encodes proteins never seen before in biology, and the remaining 40% contains very few of the fundamental metabolic genes found in almost all other eukaryotes. This indicates that the mechanism of interaction between Plasmodium and the host red blood cell may reveal novel metabolic enzymes that can provide new targets for pharmacological intervention. Using recently developed mass spectrometry techniques, we will quantitate metabolites in Plasmodium-infected cells and integrate these and other data to generate network interaction models revealing new biological insights into this deadly pathogen.

Feroz R. Papa
University of California San Francisco
Project Title: New Tools to Measure and Correct Endoplasmic Reticulum Stress in Single Living Cells
Grant ID: DP2-OD001925
Aided by chaperones and other activities, proteins of the secretory pathway fold to their correct shapes in the endoplasmic reticulum (ER). But, if ER folding capacity is exceeded, unfolded proteins start to aggregate. This imbalanced condition—called ER stress—is being linked to diverse diseases, such as type 2 diabetes and cancer. The unfolded protein response (UPR) can rebalance a stressed ER, but if the stress is too great (or the response too weak), cells appear to cross a “tipping point”, and undergo apoptosis. This could explain why overworked insulin-producing pancreatic ß-cells die in type 2 diabetes. On the other hand, a hyperactive UPR may allow malignant cells to survive hostile environments, promoting cancer. If we could measure both ER stress and the strength of corrective responses directly in healthy and diseased cells, we would be able to test these ideas quantitatively, and design rational therapies for these diseases. Structural engineers study system integrity by applying loads, measuring stresses, perturbing reinforcements, and making corrections. We propose similar approaches to ER stress disorders. Protein secretion generates and consumes oxidizing equivalents, so it should be possible to follow deviations in the ER's redox potential from its resting setpoint as an analog measure of ER stress. For this purpose, we are making redox-responsive fluorescent proteins to gauge the ER's redox potential in single, living cells. We have learned to stably perturb ER protein folding by converting an unfolded protein sensor called Ire1 into a finely adjustable rheostat. In living pancreatic ß-cells, these tools will allow us to quantify the ER stress caused by insulin folding load, in both healthy and diabetic states. Finally, using a biochemical assay we have developed, we will discover molecules targeting Ire1, to increase or decrease ER folding ability, as appropriate corrective measures for various ER stress disorders.

Dana Pe'er
Columbia University, New York Morningside
Project Title: Genetic Variation and Regulatory Networks: Mechanisms and Complexity
Grant ID: DP2-OD002414
The focus of the proposed research is to understand the effect of sequence variation on the function of molecular networks. We will develop computational algorithms that integrate genotype, gene expression and phenotype data to construct models that describe how sequence variation perturbs the regulatory network, alters signal processing and is manifested in cellular phenotypes. Our approach is based on Bayesian networks, a framework we pioneered for the reconstruction of molecular networks from high-throughput data. We recently applied this framework to develop the Geronemo algorithm which we applied to yeast and uncovered a novel relationship between the sequence specific RNA factor PUF3 and P-Bodies, as well as a Single Nucleotide Polymorphism (SNP) in MKT1 that modulates this relationship. Both novel findings were experimentally validated subsequent to their discovery. Our approach is based on the complementary duality between genetic sequence and functional genomics. A significant influence of genotype on phenotype is induced by fine tuned perturbations to the complex regulatory network that governs a cell's activity. Variation in the expression of a single gene is more tractable and can be used as an intermediary to help associate genetic factors to the more complex downstream changes in phenotype in a hierarchical fashion. Conversely, DNA sequence polymorphisms are effective perturb-agens which provide a rich source of variation to help uncover regulatory relations in the molecular network as well as direct their causality. We will develop our methods using a large collection of highly variable yeast strains, for which we have generated robust quantitative growth curves under numerous environmental conditions. The methodologies piloted in yeast will be extended to genotype and gene expression data derived from tumor samples to attempt to elucidate the multiple genetic factors that drive their proliferation. These tools will be made publicly available, including a friendly graphical user interface and visualization.

Kathrin Plath
University of California Los Angeles
Project Title: Chromatin and Epigentic Inheritance
Grant ID: DP2-OD001686
Covalent modifications of both DNA and histones are important for regulating gene expression. Here, I propose to address two fundamental, unanswered questions about chromatin modifications: i) how are changes in chromatin states established during development; and ii) once established, how are chromatin modifications stably preserved through future cell divisions? Initially, we will focus our analysis on understanding how the noncoding RNA Xist establishes silencing of the X chromosome in female mammalian cells. During initiation of X inactivation, Xist RNA spreads in cis to coat the X chromosome that will become inactive, mediates silencing, and triggers the sequential accumulation of chromatin modifications. How Xist RNA initiates silencing remains unknown. One approach to gain insight into the function of Xist is to identify interacting proteins. We have obtained results demonstrating that Xist is part of a large protein complex. In Aim 1, we therefore propose to use classical and nonconventional purification strategies to identify Xist -interacting proteins. To identify proteins necessary for maintaining the silence of the inactive X, we will perform an RNAi based screen in Aim 2, which is based on the reactivation of the inactive X. We expect to find proteins involved in the epigenetic inheritance of the silent X chromosome state. The cell cycle poses a particularly challenging problem for epigenetic inheritance since histone modifications have to be maintained when DNA strands are duplicated during S phase. A major question therefore is, how chromatin modifications are transmitted through cell divisions, and if they are indeed sufficient as carriers of the epigenetic information. Surprisingly, we have found that histone modifications on the inactive X do not accumulate throughout the cell cycle. In Aim 3, we will extend studies on the cell cycle regulation of histone modifications.

Michael P. Rape
University of California Berkeley
Project Title: Ubiquitin-Dependent Mechanisms of Tissue-Specific Cell Cycle Control
Grant ID: DP2-OD003088
This proposal describes an integrated approach to identify modules of tissue-specific cell cycle control. Despite clear evidence for tissue-specificity in proliferation and the outstanding importance of tissue-specific cell cycle regulation for development and disease, the underlying mechanisms have not been uncovered. The identification and characterization of tissue-specific modules in proliferative networks will not only provide deep insight into development of multicellular organisms and tumorigenesis, but also lay the groundwork for the discovery of tissue-specific chemotherapeutics. Because of the crucial role of ubiquitination in cell cycle control, the tissue-specific expression of ubiquitination enzymes, the frequent misregulation of ubiquitination in cancer, and the enzymatic nature of ubiquitination, we will focus the dissection of tissue-specific cell cycle regulation on members of the ubiquitination machinery. Using synthetic lethality siRNA screens in different cell lines, we will identify ubiquitination enzymes required for tissue-specific cell cycle control. By isolating substrates of these enzymes unique to a certain tissue, we will delineate pathways underlying cell cycle regulation in this tissue. Finally, by developing and miniaturizing quantifiable ubiquitination assays, we will isolate small-molecule inhibitors that allow further dissection of proliferative networks and may serve as lead structures for novel tissue-specific chemotherapeutics. We are convinced that this integrated approach using genetic discovery, biochemical dissection, and inhibitor identification based on chemical biology will provide deep insights into cell cycle and developmental regulation and lay the groundwork for novel, innovative chemotherapeutics with improved tissue-specificity.

Jody Snow Rosenblatt
University of Utah
Project Title: Identification of Signals that Extrude an Apoptotic Cell from an Epithelium
Grant ID: DP2-OD002056
I have discovered a mechanism called ‘extrusion', by which dying cells exit the epithelium without disrupting the barrier function of the layer. Here, a cell destined to die, signals its surrounding neighboring cells to form an intercellular actomyosin ring that contracts to squeeze the dying cell out. While stimuli that induce cell death (apoptosis) activate extrusion, we have also found that apoptosis and extrusion are not interdependent. This suggests that extrusion may remove cells from the epithelium in other circumstances. Signals that promote extrusion could, therefore, be used when cells leave the epithelium during developmental differentiation or initiation of tumor cell metastasis. Because most high-grade tumors have mutations in the apoptotic pathway, mutations that block apoptosis but not extrusion could enable a tumor cell to easily exit the epithelium and initiate its metastasis to other sites. Because metastasis is generally associated with cancer lethality, we believe that understanding the signalling that drives extrusion may be of utmost importance. We will also explore if extrusion might precede and induce apoptosis in overcrowded regions during normal homeostasis as a way of regulating cell numbers. Identifying the signals that trigger extrusion will be key to understanding the physiological functions that extrusion plays. We plan to identify the signalling pathway that initiates commitment of cell extrusion by investigating where this pathway bifurcates from the apoptotic pathway. We will also identify the downstream extra-cellular lipid signal that elicits extruding ring formation, as we think that this signal might provide a good molecular marker for extrusion. With these signals in hand, we will be able to test the function of extrusion in potential processes ranging from maintaining epithelial numbers and function to novel mechanisms for tumor formation and initiating tumor cell metastasis. To do so, we will extend our extrusion studies into whole zebrafish embryos.

Alan Saghatelian
Harvard University
Project Title: Discovery Metabolite Profiling of the Prolyl Peptidases
Grant ID: DP2-OD002374
Elucidation of the molecular mechanisms that underlie disease is crucial for the development of new therapeutic agents. Researchers have recently developed a number of methods to identify the genes, proteins, and metabolites associated with disease. However, complementary methods that define connections between these molecules—connections that are the foundation of biological models of disease and targeted medicine—have proven much more difficult to develop. As a result, there remains a tremendous need for innovative new approaches that reveal interactions between the molecular components of disease in vivo. The following proposal outlines the continued development and application of one such method, termed discovery metabolite profiling (DMP), for the assignment of endogenous substrates to the prolyl peptidase family of enzymes. DMP integrates an array of biological and chemical methods, including genetics, pharmacology, and analytical chemistry to identify bona fide physiological enzyme-substrate interactions. Importantly, by using DMP to study a family of enzymes that are virtually lacking in known endogenous substrates, but regulate phenotypes of tremendous biomedical interest, this research will begin to realize the incredible potential of the prolyl peptidases in medicine. Furthermore, the application of DMP to peptidases will demonstrate the generality of this approach for the future characterization of medically relevant enzymes and signaling pathways.

James Shorter
University of Pennsylvania
Project Title: Amyloid Elimination by Hsp104 and Substrate-Optimized Variants
Grant ID: DP2-OD002177
We are in dire need of innovations to combat several increasingly prevalent and inexorably lethal neurodegenerative disorders, including Alzheimer's and Parkinson's disease, which voraciously devour our social and economic resources. These disorders share a common pathogenic mechanism in which specific proteins misfold into a shared surprisingly generic conformation, termed amyloid. Amyloid fibers of diverse proteins adopt a similar ‘cross-ß' structure, in which the strands of the ß-sheets run perpendicular to the fiber axis. Further, highly cytotoxic oligomers possessing a distinct generic structure frequently accumulate prior to fibers. Although the specific protein that forms amyloid and precise localization of the deposits varies in each disease, these shared facets of amyloidogenesis bring hope that therapeutic strategies that target amyloid may have broad applications. The exceptional stability of amyloids, however, including: protease- and SDS-resistance, makes them extraordinarily difficult to clear. Indeed, they are widely perceived as intractable. Remarkably, we have found that a proteinremodeling factor from yeast, Hsp104, can rapidly disassemble amyloid fibers and oligomers comprised of the yeast prion proteins, Sup35 and Ure2. The unprecedented alacrity at which Hsp104 remodels these amyloid structures raises awareness that amyloid conformers are not intractable, and that a cellular factor capable of reversing amyloidogenesis has evolved. Curiously, although highly conserved in plants, bacteria and fungi, Hsp104 has been mysteriously lost from metazoan lineages. We hypothesize that application of Hsp104 to disease-associated amyloids may have broad therapeutic potential. Hence, we aim to: (1) annihilate amyloid fibers, oligomers and associated proteotoxicity of diverse human disease-associated proteins using Hsp104; (2) engineer or evolve substrate-optimized Hsp104 variants that attack select amyloidogenic proteins; and (3) generate Hsp104 or substrate-optimized variant therapies for animal models of amyloid-disorders. These studies will provide the foundations for new approaches to attack the devastating amyloid-disorders that plague humankind.

Dorthy A. Sipkins
University of Chicago
Project Title: Stem Cell, Tumor and Bone Marrow Microenvironment Cross-Talk In Vivo
Grant ID: DP2-OD002160
A growing body of evidence suggests that the host microenvironment plays an important role in the regulation of both normal and malignant stem cell self-renewal and differentiation. The specific locations, cellular components and molecular details of these stem cell niches are, however, little understood. Using in vivo confocal and multiphoton molecular imaging techniques to study cell interactions in the intact murine bone marrow (BM), we have defined a novel perivascular tumor and hematopoietic stem/progenitor cell (HSPC) niche. We have identified the molecules used by leukemic cells to access this niche, however the mechanisms governing HSC transit to these areas have not yet been defined. Moreover, the role of this niche in maintaining normal HSCs or coordinating specific HSC cell functions is unknown. Given the importance of HSC-based transplantation therapies as well as the clinical significance of tumor metastasis to the BM, there is great therapeutic potential in understanding these cellular interactions. The long-term goals of our research are, therefore, to define the molecular architecture and functional significance of this tumor and HSC niche using state-of-the-art in vivo imaging techniques combined with cell and molecular biology approaches. Specifically, we will 1) examine the mechanisms governing HSC transit and engraftment in these niches, 2) determine how tumor growth impacts the niche and whether tumor and benign HSCs compete within the niche, and 3) develop targeted nanoparticles to release encapsulated compounds in precise regions of the vascular niche. These nanoparticles will serve as a unique tool to elucidate the critical molecules that mediate HSC and tumor proliferation in the niche and as a prototype for a targeted drug delivery agent. Ultimately, we aim to use the knowledge from these studies to design specific anti-tumor treatments that spare normal hematopoiesis and to define factors that improve the efficacy of stem cell-based therapies.

David Spiegel
Yale University
Project Title: Small-Molecule Antibody Recruiting Therapeutics for Treating Human Disease
Grant ID: DP2-OD002913
In recent years, antibody-based therapeutics have become important instruments in treating human diseases ranging from rheumatoid arthritis to cancer. However, these approaches suffer from certain limitations including severe (often fatal) side-effects, lack of oral bioavailability, and high cost. Here, we propose an alternative method that exploits the powerful cytolytic potential of antibodies already present in the human bloodstream. We will synthesize small-molecules capable of redirecting endogenous anti-2,4-dinitrophenyl (anti-DNP) antibodies to the surfaces of various pathogenic cell-types (Figure). As shown, bifunctional molecular constructs will be composed of a bivalent antibody-binding terminus (ABT), a cell surfacebinding terminus (CBT), and a linker region. Formation of a ternary complex between these agents, anti- DNP antibodies, and target cells, will lead to targeted cytotoxicity through various mechanisms including antibody-dependent cellular cytotoxicity (ADCC), or complement-dependent cytotoxicity (CDC). Applications of this approach to cancer and HIV treatment are described, along with more general directions.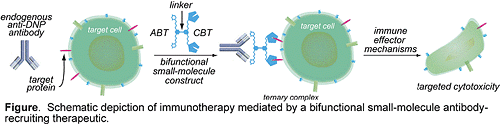 The proposed studies involve three aims: (1) to synthesize and evaluate an ABT capable of binding endogenous anti-DNP antibodies with high affinity, (2) to synthesize and evaluate a bifunctional small-molecule antiviral reagent targeting HIV gp120, and (3) to identify a small-molecule ligand for the interleukin-6 (IL-6) receptor for incorporation into bifunctional therapeutics targeting the B-cell malignancy multiple myeloma. Concise chemical syntheses of these agents are set forth, and encompass no more than six chemical transformations each. Biological evaluation will employ well established in vitro, and tissue culture models. Mathematical modeling studies are also reported that demonstrate numerically the feasibility of this approach for in vivo applications. Since high-throughput screening methods are ideally suited to identifying cell surface binding small-molecules, this general strategy is not limited to any particular type of target cell. If successful, the proposed method would represent a novel therapeutic approach to a variety of human diseases.
The proposed studies involve three aims: (1) to synthesize and evaluate an ABT capable of binding endogenous anti-DNP antibodies with high affinity, (2) to synthesize and evaluate a bifunctional small-molecule antiviral reagent targeting HIV gp120, and (3) to identify a small-molecule ligand for the interleukin-6 (IL-6) receptor for incorporation into bifunctional therapeutics targeting the B-cell malignancy multiple myeloma. Concise chemical syntheses of these agents are set forth, and encompass no more than six chemical transformations each. Biological evaluation will employ well established in vitro, and tissue culture models. Mathematical modeling studies are also reported that demonstrate numerically the feasibility of this approach for in vivo applications. Since high-throughput screening methods are ideally suited to identifying cell surface binding small-molecules, this general strategy is not limited to any particular type of target cell. If successful, the proposed method would represent a novel therapeutic approach to a variety of human diseases.

Eva M. Szigethy
University of Pittsburgh at Pittsburgh
Project Title: Understanding and Treating Neuropsychiatric Symptoms of Pediatric Physical Illness
Grant ID: DP2-OD001210
Depression is costly and has detrimental effects on disease course in physically ill populations. This proposal takes a novel multi-dimensional approach to assess the neurobiological basis of depression in chronic pediatric physical illness using inflammatory bowel disease (IBD) as a model. It also evaluates the efficacy of a modified cognitive behavioral therapy (CBT) on emotional well-being, physical health, economic costs, and neurobiological outcomes. These results will provide key building blocks for a paradigm shift within medicine by integrating behavioral health into the comprehensive medical care of physical illnesses. Little is known about how the brain and body interact to increase depressive vulnerability, particularly during key developmental periods during childhood and adolescence. Adult studies identify disruptions in limbic and prefrontal brain activity in the pathophysiology of depression. Cytokines secondary to inflammation and exogenous treatment with steroids can cause mood and cognitive changes in these same brain regions. It is important to understand the neuropsychiatric effects of IBD and its treatment on underlying brain structures during adolescence, a critical developmental period for brain maturation underlying emotional regulation and cognitive processing. More importantly, neuronal plasticity during adolescence may still allow reversibility of disease-related brain effects through teaching coping strategies for life-long illness management that could change developmental trajectories and reduce vulnerability in adulthood. Using translational neuroscience approaches, this proposal will examine: 1) brain regions that underlie emotional and cognitive processing in youth with active IBD and depression using brain functional magnetic resonance imaging compared to youth with IBD and no depression, and normal controls; and 2) the efficacy of a combined CBT-physical illness narrative intervention targeting emotional and cognitive processing compared to supportive non-directive therapy at: (a) improving emotional well being; (b) alleviating physical symptoms; and (c) reducing health care costs.

Derek Toomre
Yale University
Project Title: Novel TIRF Microscopy for Analyzing Trafficking and Signaling at the Cell Cortex
Grant ID: DP2-OD002980
My major goal is to advance knowledge about events on or near the plasma membrane. This region directly controls membrane traffic to and from the cell surface (exo- and endocytosis) and is where extracellular signals are amplified and modulated by assembly of signaling scaffolds. The introduction of total internal reflection fluorescence (TIRF) microscopy, a technique that allows unprecedented axial resolution, has revolutionized studies of dynamic processes at the cell cortex. I propose 1) to develop two highly innovative multi-angle TIRF microscopes and 2) to apply these instruments towards the elucidation of mechanisms that regulate exo- and endocytosis. These microscopes will allow the penetration depth of the light beam to be varied rapidly and avoid traditional imaging artifacts. Together with new analytical methods, they will permit high-resolution 3D imaging of a ~50- 1000 nanometer cortical region of living cells. Additionally, a highly innovative FRAP implementation will allow us to ‘pulse' photoactivate single vesicles and track their fate. I will use this novel instrumentation to expand our ongoing studies on exo- and endocytic traffic. A main new goal will be to elucidate mechanisms in the vesicular trafficking pathways that regulate levels of glucose transporters (Glut4) at the cell surface, a process whose dysfunction leads to type 2 diabetes. I will test the hypothesis that the exocyst complex participates in the spatial regulation of the insulin responsiveness of Glut4 vesicle exocytosis. Using photoactivatable Glut4-Dendra I will determine whether insulin signaling triggers a switch from lipid raft to clathrin-mediated endocytic pathways. To address where PI3K signaling acts, I will implement inducible dimerization technology to rapidly turn on/off PI(3,4,5)P3 at the plasma membrane. The innovative approaches of this proposal capitalize on my unique expertise in interdisciplinary research spanning instrumentation, cell biology, and quantitative biology and will fundamentally impact biology and a medically important field.

Jing Yang
University of California San Diego
Project Title: Epithelial-Mesenchymal Transition in Tumor Metastasis
Grant ID: DP2-OD002420
90% of cancer deaths are caused by metastatic growths in distant organs; however, the molecular basis of tumor metastasis is largely unknown. In a so-called “metastatic” primary tumor mass, only a small minority of carcinoma cells are actually migrating and on their route to disseminate into distant organs. To understand the cellular and molecular machineries that promote carcinoma cell dissemination, it is essential to identify and isolate such rare migrating carcinoma cells for molecular studies. A highly conserved developmental program Epithelial-Mesenchymal Transition (EMT) has been indicated in promoting metastasis progression. A group of embryonic EMT-inducing transcription factors, including Twist, Snail and Slug, are induced in carcinoma cells to promote cell motility and invasion. These transcription factors mainly function during embryogenesis, and they are mostly silent in normal adult tissues. Since activation of EMT is an early event to promote tumor cell dissemination, transcriptional activation of Twist, Snail and Slug provides a very specific and sensitive indicator of carcinoma cells that are undergoing EMT, migrating and disseminating into distant organs. In this study, I propose to use the promoters of Twist, Snail and Slug to drive the expression of a novel EMT detection/selection system, thus marking migrating tumor cells in vivo. Using this strategy, we aim to identify, image and isolate migrating carcinoma cells in primary tumors, to directly test the biological significance of EMT and cell migration in tumor metastasis in vivo, and to explore the signal pathways that promote carcinoma cells to migrate and disseminate into distant organs. If successful, our experiments will, for the first time, generate a molecular definition of the rare migrating carcinoma cells in vivo. The results from this study will not only significantly improve our molecular understanding of cancer metastasis, but also provide potential prognostic markers and specific targets for anti-metastasis therapies.

Mehmet F. Yanik
Massachusetts Institute of Technology
Project Title: Development of On-Chip Ultra High-Throughput Whole-Animal Assay Technologies
Grant ID: DP2-OD002989
In recent years, the advantages of using the nematode Caenorhabditis elegans as a model system for human disease have become increasingly apparent, culminating in two Nobel Prizes in Physiology and Medicine within the last five years. Existing vertebrate animal models, and the instrumentation incorporated to study them, cannot be utilized for high throughput assays for rapid identification of novel genes, and drug leads. The model organism C. elegans allows in vivo genome-wide assays and high-throughput screens due to the availability of unmatched genetic techniques, its transparency, and the ability to grow it in minute volumes. Yet, since the first publications in early 1960s, little has changed how scientists manipulate this tiny organism manually, as a result of which even simple large-scale assays still take months to years to complete. Importantly, due to the lack of key technologies, several assays either cannot be performed at all or have to be dramatically simplified for high-throughput screens. We propose (1) development of the first on-chip ultra high-throughput whole-animal manipulation technologies for in vivo drug screens and genome-wide discovery of gene functions and interactions by complex on-chip behavioral and biochemical assay strategies; and (2) the first large-scale in vivo study of mechanisms of neural degeneration and regeneration following reproducible injuries using the proposed techniques. The proposed technologies and high-throughput assay strategies can significantly impact discovery of many molecular mechanisms using disease models of small animals.



