Science Highlights Archives
No Two Tumors are Quite Alike: Metabolomics Reveals Breast Cancers' Different Energy Needs
 Research from Metabolomics Program awardee Dr. Susan Sumner from the University of North Carolina showed that breast cancer cells from different tumor types use energy in different ways. In the study, Dr. Sumner and colleagues used a novel experimental approach to investigate metabolomics - the study of molecules produced and consumed by our metabolisms. The researchers investigated two types of breast cancer tumor subtypes. The first type, called luminal A subtype, is found in approximately 30-70% of breast cancer cases and tends to have the best chances for patients to recover. The second type, called the triple negative subtype, accounts for roughly 15-20% of breast cancer cases and is more difficult to treat. It is important for scientists to figure out how the triple negative subtype is different from easier-to-treat subtypes so better remedies can be developed in the future.
Research from Metabolomics Program awardee Dr. Susan Sumner from the University of North Carolina showed that breast cancer cells from different tumor types use energy in different ways. In the study, Dr. Sumner and colleagues used a novel experimental approach to investigate metabolomics - the study of molecules produced and consumed by our metabolisms. The researchers investigated two types of breast cancer tumor subtypes. The first type, called luminal A subtype, is found in approximately 30-70% of breast cancer cases and tends to have the best chances for patients to recover. The second type, called the triple negative subtype, accounts for roughly 15-20% of breast cancer cases and is more difficult to treat. It is important for scientists to figure out how the triple negative subtype is different from easier-to-treat subtypes so better remedies can be developed in the future.
The researchers used advanced metabolomics methods to incorporate special labels into the molecules of the cancer cells. They observed that the triple negative cancer cells consumed more of the molecule glutamine, but less glucose than the luminal A cells. Glucose is a major energy source for cells, including cancer cells. Some cells can switch to less efficient energy sources, like glutamine, when energy is scarce. This finding may help us understand how triple negative forms of breast cancer can survive better than luminal A subtypes in response to cancer treatments, which often reduce the amount of energy cancer cells can use. By adapting to low energy environments, triple negative cancer cells can live despite the survival pressures resulting from treatment. Understanding how different cancer subtypes use energy may allow researchers to develop more targeted therapeutics for even the rarest and most difficult-to-treat cancer types. Additionally, the metabolomics methods used by the Sumner research team may be useful to study the differences in tumor subtypes in cancers other than breast cancer, potentially leading to more targeted methods for fighting different tumors in individual patients.
Reference: Stable Isotope-Resolved Metabolomic Differences between Hormone-Responsive and Triple-Negative Breast Cancer Cell Lines. Winnike JK, Stewart DA, Pathmasiri WW, McRitchie SL, Sumner SJ. Int J Breast Cancer, vol 2018 2063540. 30 Sep. 2018, doi:10.1155/2018/2063540
Energy Use Monitored in Cancer Cells with New Breakthrough Technique
 Metabolomics is the study of the small molecules consumed and produced by our metabolism. One of the benefits of metabolomics research is that it provides information about how the cells in our bodies break down molecules to produce energy. This is particularly important in cancer, as cancer cells often use unusual methods of energy production that are distinct from those used by non-cancer cells. One common way for researchers to study this is to introduce labeled molecules into different experimental systems, such as research animals. The labeled molecules are easier to tell apart from the other molecules that make up our metabolism and can be monitored to see how they are broken down and incorporated into new biomolecules. However, these animal experiments are not optimal, as restraining the animals can induce stress and the experiments often require sedation. Both stress and sedation can alter metabolic processes. Moreover, these methods are invasive, and so they are only used over a short timeframe, which sets a limit on the information they provide.
Metabolomics is the study of the small molecules consumed and produced by our metabolism. One of the benefits of metabolomics research is that it provides information about how the cells in our bodies break down molecules to produce energy. This is particularly important in cancer, as cancer cells often use unusual methods of energy production that are distinct from those used by non-cancer cells. One common way for researchers to study this is to introduce labeled molecules into different experimental systems, such as research animals. The labeled molecules are easier to tell apart from the other molecules that make up our metabolism and can be monitored to see how they are broken down and incorporated into new biomolecules. However, these animal experiments are not optimal, as restraining the animals can induce stress and the experiments often require sedation. Both stress and sedation can alter metabolic processes. Moreover, these methods are invasive, and so they are only used over a short timeframe, which sets a limit on the information they provide.
The Resource Center for Stable Isotope-Resolved Metabolomics (RC-SIRM) at the University of Kentucky, funded by the Common Fund Metabolomics Program, recently devised a new method of monitoring metabolic processes that avoids these concerns. In the new protocol, researchers feed labeled molecules to animals using a liquid diet, which is noninvasive and does not require that the animal be restrained or anaesthetized. This results in less stress for the animal, which leads to more accurate experimental outcomes. This method also allows for a longer timeframe of experimentation, letting researchers follow the metabolism of the labeled molecules through long metabolic pathways. Using this new method, RC-SIRM was able to show that human lung cancer cells implanted in mice demonstrate a distinct metabolism. Some of the changes include reduced glycolysis – or breakdown of sugars – and more active synthesis of glycogen, an energy storage molecule. These changes allow tumor cells to live in energy deficient environments. Metabolic adaptations of human tumor cells, like the ones described here, could have important implications for a tumor cell’s ability to survive in the body. These findings were only possible because of the new liquid diet method, an effective and noninvasive solution that allows researchers to study how cells alter their metabolism to promote growth and survival.
Reference: Noninvasive liquid diet delivery of stable isotopes into mouse models for deep metabolic network tracing. Ramon C. Sun, Teresa W.-M. Fan, Pan Deng, Richard M. Higashi, Andrew N. Lane, Anh-Thu Le, Timothy L. Scott, Qiushi Sun, Marc O. Warmoes, and Ye Yang. Nature communications, November, 2017, doi: 10.1038/s41467-017-01518-z
View the video below to also learn about this exciting research!
Untangling Metabolomic Pathways: How do Individual Traits Impact Results?

Metabolomics is a powerful research approach that allows scientists to identify specific chemicals associated with many different biological processes occurring in the human body. Often, the levels of these chemicals change in response to disease. Researchers try to find these changes, known as biomarkers, early in disease for diagnostic purposes and to track disease progression. Metabolomics methods allow scientists and doctors to test for these biomarkers. However, individual traits and lifestyle factors, such as diet, smoking, age, and exercise, can make it difficult to interpret these metabolic changes.
Metabolomics program researcher Dr. Min Zhang of Purdue University, contributed to a recent study to account for different individual traits when searching for biomarkers using metabolomics-based experiments. Dr. Zhang and her colleagues used a statistical model called seemingly unrelated regression (SUR) to account for the effects of individual factors when comparing samples from colorectal cancer patients and healthy controls. Using the SUR method, the research team teased out biomarkers unique to the colorectal cancer patients. In addition, they placed these biomarkers into biological pathways, potentially allowing researchers to identify further differences in patients with disease. These results are an exciting advancement for the identification of unique biomarkers to help with disease diagnosis using metabolomic methods. Additionally, they highlight the importance of taking an individual’s history into account when assessing chemical changes that occur in the disease state.
Reference:
Altered metabolite levels and correlations in patients with colorectal cancer and polyps detected by using seemingly unrelated regression analysis. Chen C, Nagana Gowda GA, Zhu J, Deng L, Gu H, Gabreila Chiorean E, Abu Zaid M, Harrison M, Zhang D, Zhang M, Raftery D. Metabolism. 2017, 13:125.
Note: article full text may require institutional access
Metabolomics Study Helps Explain How Physical Activity Improves Muscle Health
A recent study from Metabolomics awardee K. Skreekumaran Nair and colleagues at the Mayo Clinic in Rochester, Minnesota has produced exciting information about how different exercise routines can improve muscle health in old and young adults. It is well known that physical activity can benefit health, but we still have a lot to learn about the biological changes that lead to this improvement. Research supported by the Common Fund’s Metabolomics program has the potential to enhance our understanding of how physical activity betters our health.

The study from Dr. Nair and colleagues examined how three different exercise routines impacted levels of proteins that influence muscle health in old and young adults. The three exercise routines were high-intensity aerobic interval training (repeating short bouts of activity at near-maximal intensity), resistance training (lifting weights or bands), and combination training (combination of both methods, although the intensities of the aerobic or resistance components are less than the individual methods).
Using multiple metabolomics and proteomics methods, the study showed that all three exercise routines increased protein synthesis in muscle cells. Increases included elevated production of proteins involved in the formation of new blood vessels, further elevation of protein synthesis levels, and energy production in the mitochondria. These changes led to an improvement in muscle function by making the muscles stronger and providing more energy to the muscles during and after exercise. While all exercise methods showed health benefits, the high-intensity aerobic interval training had the most pronounced effect. Importantly, they also found that these exercise routines could reverse age-related decline in muscle function in older adults. This exciting research is an important step in our understanding how exercise can improve our health, and may help future studies design personalized exercise routines to help combat disease and age-related health issues.
Reference:
Enhanced Protein Translation Underlies Improved Metabolic and Physical Adaptations to Different Exercise Training Modes in Young and Old Humans. Robinson MM, Dasari S, Konopka AR, Johnson ML, Manjunatha S, Esponda RR, Carter RE, Lanza IR, Nair KS. Cell Metabolism, 2017, March 7; 25(3):581-592
New Metabolomics Software Improves Data Analysis and Integration
 Metabolomics is the study of all the small molecules, called metabolites, that are produced or consumed during the chemical reactions that sustain life. The study of metabolites is very important, as it can show early warning signs of diseases and provide information about how efficiently the cells of our body are working. Metabolomics is a maturing field, and a goal of the NIH Common Fund’s Metabolomics program is to support the development of new tools to better understand, analyze, and use metabolomics data.
Metabolomics is the study of all the small molecules, called metabolites, that are produced or consumed during the chemical reactions that sustain life. The study of metabolites is very important, as it can show early warning signs of diseases and provide information about how efficiently the cells of our body are working. Metabolomics is a maturing field, and a goal of the NIH Common Fund’s Metabolomics program is to support the development of new tools to better understand, analyze, and use metabolomics data.
Metabolomics program grantee Dr. Oliver Fiehn and collaborators have developed a new metabolomics software tool, called Metabox, to improve the analysis of metabolomics data and its integration with other biological information. This free, web-based software allows scientists to easily visualize metabolic networks. Software tools like Metabox are an important step towards improving our ability to connect metabolomics findings with other biological information, and sets the stage for future groundbreaking discoveries across many fields.
Reference:
Metabox: A Toolbox for Metabolomic Data Analysis, Interpretation, and Integrative Exploration. Wanichthanarak K, Fan S, Grapov D, Barupal DK, and Fiehn O. PLOS One, 2017, Jan 31;12(1)
Metabolomics Study Highlights Potential Early Warning Signs for Diabetes-Related Complications
 Diabetes is a leading cause of illness across the world, and awardees looking at diabetes are supported by several Common Fund programs. Both Type I and Type II diabetes are linked to changes in metabolism, such as the way our bodies process sugars or fats. In addition, diabetes-related complications can affect other tissues of the body, such as the kidney, the peripheral nerves, or the retina, also showing changes in metabolism. Researchers supported by the Common Fund’s Metabolomics program are interested in learning more about how altered metabolism contributes to diabetes and diabetes-related complications.
Diabetes is a leading cause of illness across the world, and awardees looking at diabetes are supported by several Common Fund programs. Both Type I and Type II diabetes are linked to changes in metabolism, such as the way our bodies process sugars or fats. In addition, diabetes-related complications can affect other tissues of the body, such as the kidney, the peripheral nerves, or the retina, also showing changes in metabolism. Researchers supported by the Common Fund’s Metabolomics program are interested in learning more about how altered metabolism contributes to diabetes and diabetes-related complications.
A recent study from Common Fund’s Metabolomics program grantee Dr. Charles Burant and colleagues has shown that there are tissue-specific metabolic changes following the onset of diabetes in both mice and humans. For example, the research showed that levels of particular metabolites were increased specifically in the urine, and not in other tissues or the blood. This suggests that tissue-specific metabolic changes may provide key information about diabetic complications and suggests that urine biomarkers may be most effective for tracking diabetic kidney disease progression.
Reference:
Tissue-specific Metabolic Reprogramming Drives Nutrient Flux in Diabetic Complications. Sas KM, Kayampilly P, Byun J, Nair V, Hinder LM, Hur J, Zhang H, Lin C, Qi NR, Michailidis G, Groop PH, Nelson RG, Darshi M, Sharma K, Schelling JR, Sedor JR, Pop-Bosui R, Weinberg JM, Soleimanpour SA, Abcouwer SF, Gardner TW, Burant CF, Feldman EL, Kretzler M, Brosius FC 3rd, and Pennathur S. Journal of Clinical Investigation Insight, 2016, Sep 22;1(15)
Metabolomics Reveals New Cross-Talk Between Tumors and Surrounding Cells
 Tumors are rapidly dividing group of cells which require significant amounts of nutrients and energy to continue to grow. Understanding both the requirements and sources of nutrients needed by tumors can be a key step in our effort to treat and prevent cancer. A recent collaboration led by Dr. Alec Kimmelman from Harvard University enlisted the help of the Michigan Regional Comprehensive Metabolomics Research Core, supported by the NIH Common Fund’s Metabolomics program, to generate groundbreaking evidence about how one type of cancer cell hijacks nearby cells to supply it with energy. Researchers found that tumor cells in a type of pancreatic cancer can use a particular small chemical messenger (metabolite) obtained from surrounding cells as a source of energy to promote growth of the tumor.
Tumors are rapidly dividing group of cells which require significant amounts of nutrients and energy to continue to grow. Understanding both the requirements and sources of nutrients needed by tumors can be a key step in our effort to treat and prevent cancer. A recent collaboration led by Dr. Alec Kimmelman from Harvard University enlisted the help of the Michigan Regional Comprehensive Metabolomics Research Core, supported by the NIH Common Fund’s Metabolomics program, to generate groundbreaking evidence about how one type of cancer cell hijacks nearby cells to supply it with energy. Researchers found that tumor cells in a type of pancreatic cancer can use a particular small chemical messenger (metabolite) obtained from surrounding cells as a source of energy to promote growth of the tumor.
In order to continue growing, pancreatic ductal adenocarcinoma tumor cells have figured out how to send signals to neighboring cells to give them energy through a cellular process called autophagy. During this process, the surrounding cells break down their own components into smaller metabolites, which are then released for the tumor cells to use as energy to survive and grow. This previously unknown cross-talk between tumor cells and their neighbors is an exciting find, as it helps us better understand how tumors continue to grow even in nutrient-poor conditions. The report also highlights a potential new target for cancer therapies, showing that if you stop the nearby cells from releasing their metabolites, the tumor cells stop growing. This research illustrates the power of metabolomics to provide the scientific community with exciting tools and data to better understand complex diseases.
Reference:
Pancreatic stellate cells support tumour metabolism through autophagic alanine secretion. Sousa CM, Biancur DE, Wang X, Halbrook CJ, Sherman MH, Zhang L, Kremer D, Hwang RF, Witkiewicz AK, Ying H, Asara JM, Evans RM, Cantley LC, Lyssiotis CA, Kimmelman AC. Nature. 2016 Aug 25;536(7617):479-83.
Common Fund Supports Training for Incorporation of Metabolomics into Research Studies
One of the goals of the Common Fund Metabolomics program is to provide the training necessary for metabolomics techniques to be widely adopted by the biomedical research community. As a part of that goal, the Common Fund has sponsored an annual four-day metabolomics workshop led by Dr. Stephen Barnes at the University of Alabama at Birmingham. The workshop includes education and hands-on training in the entirety of a metabolomics experiment, from designing the experiment and collecting the samples to running the instrumentation, analyzing the data, and interpreting the results.
Dr. Barnes and colleagues have summarized the training they offer in these workshops in two papers published in the Journal of Mass Spectrometry. The first paper in the series focuses on the principles and practices needed to design an effective metabolomics experiment and the variety of techniques and instrumentation used to produce the raw data. The second paper then describes the analysis and interpretation of the data produced as well as some thoughts about the future of metabolomics.
Learn more about the UAB metabolomics workshops here.
References:
Training in metabolomics research. I. Designing the experiment, collecting and extracting samples and generating metabolomics data. Barnes S, Benton HP, Casazza K, Cooper SJ, Cui X, Du X, Engler JA, Kabarowski JH, Li S, Pathmasiri W, Prasain JK, Renfrow MB, Tiwari HK. J Mass Spectrom. 2016 Jul; 51(7) 461-75.
Training in metabolomics research. II. Processing and statistical analysis of metabolomics data, metabolite identification, pathway analysis, applications of metabolomics and its future. Barnes S, Benton HP, Casazza K, Cooper SJ, Cui X, Du X, Engler JA, Kabarowski JH, Li S, Pathmasiri W, Prasain JK, Renfrow MB, Tiwari HK. J Mass Spectrom. 2016 Aug; 51(8) 535-48.
Antibiotics Treatment Leads to Harmful Infection by Decreasing Key Metabolites
 The use of antibiotics to treat acute infections can have harmful side effects, such as colonization of the intestine by the harmful bacterium Clostridium difficile. The Common Fund’s Metabolomics Program grantee Dr. Casey Theriot and colleagues have helped uncover the mechanism by which antibiotic treatment causes increased susceptibility to Clostridium difficile infection (CDI). Dr. Theriot hypothesized that this antibiotic effect occurs because bacteria killed by the antibiotics normally produce a metabolite that prevents CDI from developing. To test this hypothesis, the researchers monitored the levels of key intestinal metabolites called bile acids in the intestines of mice before and after treatment with a variety of antibiotics. Using this information, as well as testing whether the antibiotics altered the ability of Clostridium difficile to grow in the intestines of these mice, they were able to determine the amount of specific bile acids required to prevent CDI. Finally, they confirmed their findings by adding back the lost bile acids after antibiotic treatment. This information should be key to helping design therapeutics that could prevent CDI in the future.
The use of antibiotics to treat acute infections can have harmful side effects, such as colonization of the intestine by the harmful bacterium Clostridium difficile. The Common Fund’s Metabolomics Program grantee Dr. Casey Theriot and colleagues have helped uncover the mechanism by which antibiotic treatment causes increased susceptibility to Clostridium difficile infection (CDI). Dr. Theriot hypothesized that this antibiotic effect occurs because bacteria killed by the antibiotics normally produce a metabolite that prevents CDI from developing. To test this hypothesis, the researchers monitored the levels of key intestinal metabolites called bile acids in the intestines of mice before and after treatment with a variety of antibiotics. Using this information, as well as testing whether the antibiotics altered the ability of Clostridium difficile to grow in the intestines of these mice, they were able to determine the amount of specific bile acids required to prevent CDI. Finally, they confirmed their findings by adding back the lost bile acids after antibiotic treatment. This information should be key to helping design therapeutics that could prevent CDI in the future.
Read the press release accompanying this publication. ![]()
Reference:
Antibiotic-Induced Alterations of the Gut Microbiota Alter Secondary Bile Acid Production and Allow for Clostridium difficile Spore Germination and Outgrowth in the Large Intestine. Theriot CM, Bowman AA, Young YB. mSphere. 2016 Jan 6; 1(1):eooo45-15.
Metabolomics Scientists Tie Metabolites to Lifespan in Mammals
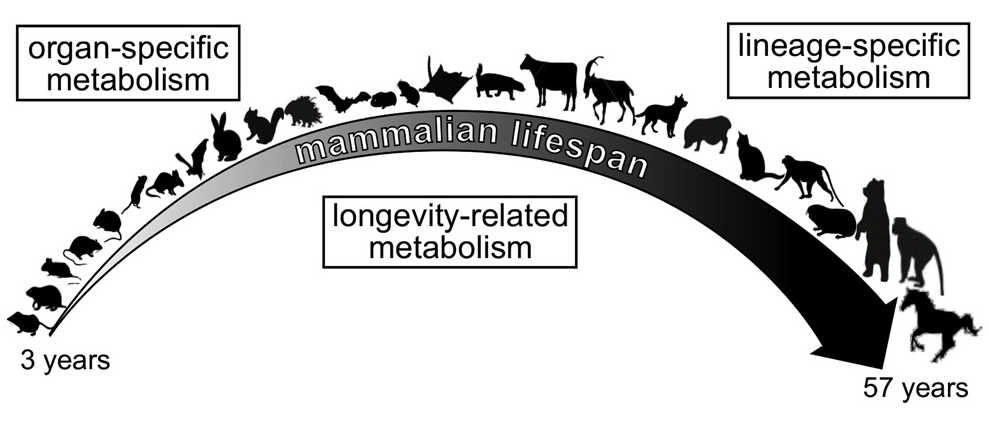 Despite sharing a common ancestor, mammals have evolved to have a wide range of lifespans. These varying lifespans require adjustment to many aspects of mammalian biology, including metabolic rates. A research team including a grantee from the NIH Common Fund’s Metabolomics program has investigated the association of metabolism with lifespan by measuring the levels of over two hundred metabolites in the brains, hearts, kidneys, and livers of 26 diverse species of mammals. Dr. Sun Hee Yim and colleagues discovered that while each species and each organ had a distinct pattern of metabolites, the amounts of certain metabolites correlated with species lifespan. They were also able to confirm many of their findings in mice that were chemically or genetically manipulated to have extended lifespans. Further research will be required to determine whether the changes in metabolites contribute to extending the lifespan or instead result from having an extended lifespan.
Despite sharing a common ancestor, mammals have evolved to have a wide range of lifespans. These varying lifespans require adjustment to many aspects of mammalian biology, including metabolic rates. A research team including a grantee from the NIH Common Fund’s Metabolomics program has investigated the association of metabolism with lifespan by measuring the levels of over two hundred metabolites in the brains, hearts, kidneys, and livers of 26 diverse species of mammals. Dr. Sun Hee Yim and colleagues discovered that while each species and each organ had a distinct pattern of metabolites, the amounts of certain metabolites correlated with species lifespan. They were also able to confirm many of their findings in mice that were chemically or genetically manipulated to have extended lifespans. Further research will be required to determine whether the changes in metabolites contribute to extending the lifespan or instead result from having an extended lifespan.
Reference:
Organization of the Mammalian Metabolome according to Organ Function, Lineage Specialization, and Longevity. Ma S, Yim SH, Lee SG, Kim EB, Lee SR, Chang KT, Buffenstein R, Lewis KN, Park TJ, Miller RA, Clish CB, Gladyshev VN. Cell Metab. 2015 Aug 4; 22(2):332-43.
Features of the Metabolomics Workbench highlighted in a recent publication
 The Metabolomics Program’s Data Repository and Coordination Center website, the Metabolomics Workbench, is the subject of a recent publication in Nucleic Acids Research. Housed at the San Diego Supercomputer Center (SDSC), the Metabolomics Workbench serves as a home for all of the resources generated by the Common Fund Metabolomics program. The website is meant to fill a need in the metabolomics community for a centralized data repository in which the research public can both upload their own data and download existing datasets for additional analysis. It is also a portal for access to metabolite standards, metabolomics structures, protocols, data analysis and visualization tools, and training materials generated by the Metabolomics program. The research article focuses on the functionality of the site, the details of the information housed at site, and the underlying computational structure that runs the site.
The Metabolomics Program’s Data Repository and Coordination Center website, the Metabolomics Workbench, is the subject of a recent publication in Nucleic Acids Research. Housed at the San Diego Supercomputer Center (SDSC), the Metabolomics Workbench serves as a home for all of the resources generated by the Common Fund Metabolomics program. The website is meant to fill a need in the metabolomics community for a centralized data repository in which the research public can both upload their own data and download existing datasets for additional analysis. It is also a portal for access to metabolite standards, metabolomics structures, protocols, data analysis and visualization tools, and training materials generated by the Metabolomics program. The research article focuses on the functionality of the site, the details of the information housed at site, and the underlying computational structure that runs the site.
Metabolomics Workbench: An international repository for metabolomics data and metadata, metabolite standards, protocols, tutorials and training, and analysis tools. Manish Sud1, Eoin Fahy, Dawn Cotter, Kenan Azam, Ilango Vadivelu, Charles Burant, Arthur Edison, Oliver Fiehn, Richard Higashi, K. Sreekumaran Nair, Susan Sumner, and Shankar Subramaniam. Nucleic Acids Research. 2015.
Taming the Torrent of Metabolic Data
 Clinical tests that identify and measure levels of biological molecules in biofluids are now commonplace. Doctors routinely measure levels of molecules like blood glucose or cholesterol. These tests provide valuable information about the state of a patient’s health, but imagine what could be accomplished if your doctor could measure hundreds of biological molecules from a single blood draw. Metabolomics could make that possible!
Clinical tests that identify and measure levels of biological molecules in biofluids are now commonplace. Doctors routinely measure levels of molecules like blood glucose or cholesterol. These tests provide valuable information about the state of a patient’s health, but imagine what could be accomplished if your doctor could measure hundreds of biological molecules from a single blood draw. Metabolomics could make that possible!
Metabolomics is a field of biomedical science that provides a snapshot of metabolites – the molecules formed and broken in the chemical reactions that sustain life. It has the potential to uncover new biomarkers for health and disease and to contribute to non-invasive diagnostics. However, the sheer number of metabolites participating in metabolism at any given time makes analyzing metabolomics data challenging. One of the biggest challenges is to be able to identify the molecules detected through untargeted metabolomics experiments.
Common Fund Metabolomics Program grantee, Dr. Oliver Fiehn at the NIH West Coast Metabolomics Center at UC Davis, and his colleagues in Japan, have developed open source software called MS-DIAL to help researchers analyze their untargeted metabolomics data. MS-DIAL uses mathematical algorithms to convert the torrent of data generated by a technique called mass spectrometry into information about the different metabolites present. It then looks for a match between chemical properties of the metabolites and known molecules already included in publically available molecular databases. Through this process, MS-DIAL enables efficient molecule identification as well as uncovering information about the amount of particular molecules present in the biological samples. Dr. Fiehn and his colleagues put MS-DIAL to the test by applying it to nine different samples of algae. Algae are relatively simple to culture in the lab, but they are metabolically complex, and serve as a good test subject for metabolomics techniques. The researchers were able to identify 1023 different lipid molecules from the algae, including some lipids that had never been identified before! The hope is that the MS-DIAL software will help other researchers squeeze as much information out of every metabolomics experiment as possible.
MS-DIAL: data-independent MS/MS deconvolution for comprehensive metabolome analysis. Tsugawa H, Cajka T, Kind T, Ma Y, Higgins B, Ikeda K, Kanazawa M, VanderGheynst J, Fiehn O, Arita M. June 2015. Nature Methods;12(6): 523-6.
Study Finds Potential New Drug Target for Lung Cancer
 Scientists have long known that the metabolism of tumor cells differs from normal, healthy cells. However, it has been challenging to study tumor metabolism in living tumor cells from a large number of cancer patients. Metabolomics, a technique that allows scientists to analyze the molecules involved in metabolism, may change that.
Scientists have long known that the metabolism of tumor cells differs from normal, healthy cells. However, it has been challenging to study tumor metabolism in living tumor cells from a large number of cancer patients. Metabolomics, a technique that allows scientists to analyze the molecules involved in metabolism, may change that.
Researchers at the Common Fund-supported Resource Center for Stable Isotope-Resolved Metabolomics (RC-SIRM) looked at how the molecule glucose is broken down as part of the metabolism of patients with non-small-cell lung cancer. Shortly before surgery for tumor removal, the researchers injected patients with a form of glucose that is non-radioactively labeled. Using metabolomic techniques, researchers can follow how the labeled glucose is broken down by both the tumor cells and healthy cells from the same patient. As the glucose is broken down into component molecules, the label will remain with one or more of the components. These labeled components are a clue to which metabolic processes are active in the tumor.
The RC-SIRM team found that tumor cells broke down the glucose using two processes, glycolysis and the tricarboxylic acid cycle (TCA cycle). They also found that an enzyme involved in the TCA cycle, called pyruvate carboxylase, was produced in greater quantities in tumors compared to healthy tissue. Reducing the level of this enzyme in lab-grown human lung cancer cells reduced cell growth and interfered with the ability of these cells to form tumors in mice. This study by the RC-SIRM team demonstrates the exciting potential for metabolomic approaches to reveal new pathways and players in cancer metabolism that may become novel targets for cancer therapy and diagnostics.
RC-SIRM at the University of Kentucky is a Regional Metabolomics Resource Core funded through the Common Fund Metabolomics program. Learn more about the Metabolomics program.
The original research article was published in the Journal of Clinical Investigation. Read the article abstract.
Using metabolomics to study cancer cell metabolism
 Cancer cells have distinctive changes in their metabolism that can be exploited for cancer diagnosis and treatment. One metabolic change that occurs is in the way a cancer cell uses the biological molecule glutamine. This chemical is both broken down for energy and used as a starting point in the synthesis of other biological compounds such as nucleotides and amino acids. Drugs that inhibit glutamine processing by targeting the enzyme glutaminase, which converts glutamine into glutamate, may be promising for cancer therapy. However, these therapies would be most useful if we could easily identify the patients who are mostly likely to benefit from them. Towards this end, a recent study from the West Coast Metabolomics Center examined the glutamate to glutamine ratio (GGR) in breast tissue from 270 breast cancer patients compared to 97 normal controls. They found that this ratio was significantly higher in cancer tissue. Tumor characteristics such as estrogen receptor (ER) status and tumor grade correlated with GGR, such that ER negative breast tumors and higher grade tumors had more elevated GGR levels. The finding that GGR levels are elevated in many breast tumors suggest that this measurement might predict which tumors would be candidates for treatment with newly developed glutaminase inhibitors.
Cancer cells have distinctive changes in their metabolism that can be exploited for cancer diagnosis and treatment. One metabolic change that occurs is in the way a cancer cell uses the biological molecule glutamine. This chemical is both broken down for energy and used as a starting point in the synthesis of other biological compounds such as nucleotides and amino acids. Drugs that inhibit glutamine processing by targeting the enzyme glutaminase, which converts glutamine into glutamate, may be promising for cancer therapy. However, these therapies would be most useful if we could easily identify the patients who are mostly likely to benefit from them. Towards this end, a recent study from the West Coast Metabolomics Center examined the glutamate to glutamine ratio (GGR) in breast tissue from 270 breast cancer patients compared to 97 normal controls. They found that this ratio was significantly higher in cancer tissue. Tumor characteristics such as estrogen receptor (ER) status and tumor grade correlated with GGR, such that ER negative breast tumors and higher grade tumors had more elevated GGR levels. The finding that GGR levels are elevated in many breast tumors suggest that this measurement might predict which tumors would be candidates for treatment with newly developed glutaminase inhibitors.
The unique metabolism of bacteria that colonize chronic wounds
 Infection with bacterial communities contributes to the conversion of an acute wound to a chronic state. Using Nuclear Magnetic Resonance Spectrometry, Dr. Ammons and her colleagues looked at the metabolite composition of Staphylococcus aureus bacteria from two classes: Drug-resistant highly virulent bacteria obtained from clinical isolates and non-virulent drug-sensitive lab strains. For each strain, they analyzed samples grown in standard liquid cultures and others grown under biofilm conditions that mimic the chronic wound environment. They found that both bacterial strains exhibited distinct metabolite profiles when grown in the wound-like environment. These results suggest the possibility of developing molecular markers that could be used to classify the bacteria found within a wound and determine conversion to a chronic wound state. The nature of the metabolites also provide clues to the biological changes that occur when bacteria are grown in chronic wound conditions, offering the potential to develop therapeutic agents that exploit their distinct metabolism. View the article abstract here.
Infection with bacterial communities contributes to the conversion of an acute wound to a chronic state. Using Nuclear Magnetic Resonance Spectrometry, Dr. Ammons and her colleagues looked at the metabolite composition of Staphylococcus aureus bacteria from two classes: Drug-resistant highly virulent bacteria obtained from clinical isolates and non-virulent drug-sensitive lab strains. For each strain, they analyzed samples grown in standard liquid cultures and others grown under biofilm conditions that mimic the chronic wound environment. They found that both bacterial strains exhibited distinct metabolite profiles when grown in the wound-like environment. These results suggest the possibility of developing molecular markers that could be used to classify the bacteria found within a wound and determine conversion to a chronic wound state. The nature of the metabolites also provide clues to the biological changes that occur when bacteria are grown in chronic wound conditions, offering the potential to develop therapeutic agents that exploit their distinct metabolism. View the article abstract here.
Click on the image to view Dr. Ammons' video contest submission
that explains her research in plain language with the help of some animated bacteria!
Studying the ebb and flow of biological molecules in the metabolism with metabolomics
Dr. Gary Patti and his team at the Washington University School of Medicine have built on recent advances in the fields of metabolomics and bioinformatics to develop a new approach to studying the ebb and flow of biological molecules as they are processed during metabolism. Their approach combines two techniques called “untargeted metabolomics analysis” and “isotopic labeling” that allow them to track the fate of a biological molecule in an unbiased manner. This publication describes their unique experimental process and introduces a new software program they’ve developed to make data analysis easier. They describe a test case which experimentally validates their system and demonstrates that it can be useful in identifying new biochemical pathways. Their system also allows us follow the fate of individual biological molecules in response to environmental changes. Read the article abstract here.
MetabolomeXchange: the beginnings of international metabolomics data sharing
The NIH Common Fund Metabolomics program contributes to international efforts to make metabolomics datasets accessible to researchers world-wide through the metabolomeXchange. Data from the program's Metabolomics Workbench is searchable through the metabolomeXchange website which also has links to the deposited data.
Using metabolomics to understand the interplay of antibiotics and a pathogenic bacterium that can live in the gut
Metabolomics Program Mentored Research Scientist Dr. Casey Theriot, using the Michigan Regional Comprehensive Metabolomics Resource Center (MRC2 ), published in Nature Communications that metabolites produced by gut microbes change in response to antibiotic treatment and favor growth of the pathogenic bacterium C. difficile.
Researchers look at the relationship between metabolic health and the metabolites found in blood
New work from the NIH West Coast Metabolomics Center at UC Davis describes how the metabolites found in the blood of women who are obese, sedentary and insulin-resistant change after the women participate in a weight loss and exercise intervention. Some of the metabolites that changed were derived from the gut, and may have originated either from diet or from gut-dwelling microbes. By revealing an association between improved metabolic health and an altered metabolite profile, this study and others like it could help us understand, diagnose and treat complex metabolic disorders like type 2 diabetes mellitus. Click here to access the research article.



