 Science Highlights Archive
Science Highlights Archive
Hundreds of New Genes Linked to Blindness and Other Vision Disorders
 Researchers from the Knockout Mouse Phenotyping Program (KOMP2) (part of the International Mouse Phenotyping Consortium (IMPC)) have discovered new genes linked to blindness and other vision disorders in mice. Some of these genes may also be important in human eye development and vision, offering hope of increased understanding of vision disorders and ultimately new treatments for individuals with hereditary eye disease.
Researchers from the Knockout Mouse Phenotyping Program (KOMP2) (part of the International Mouse Phenotyping Consortium (IMPC)) have discovered new genes linked to blindness and other vision disorders in mice. Some of these genes may also be important in human eye development and vision, offering hope of increased understanding of vision disorders and ultimately new treatments for individuals with hereditary eye disease.
Understanding the genetic basis of eye diseases is a major challenge. Researchers have studied the histories of families affected by eye disorders and had previously identified numerous genes associated with a wide array of eye disease. However, this type of analysis is limited and slow, as it requires following multiple generations of people over time. Other small-scale studies with laboratory animals only investigate a single gene or handful of genes. These studies are informative, but not comprehensive. The method used by the KOMP2 researchers uses large numbers of "knockout mice" - mice that lack a single specific gene – that are carefully examined for many potential biological effects, including vision impairments. With this high throughput method, large numbers of individual genes can be investigated to establish whether the missing gene is important to eye function.
In this study, the researchers evaluated 4,364 genes. Of these genes, 347 were identified as affecting vision and 75% (261) of genes affecting vision were linked to eye function and pathology for the first time. Overall, the genes and gene families revealed in this study will serve as a powerful resource for geneticists who can now scan whole genome sequencing data for patients whose vision disorders are not linked to any of the previously known genes affecting vision. In the future, these mouse knockout models could also be used to test targeted therapies, including potential medications or even gene therapies.
Reference:
Identification of genes required for eye development by high-throughput screening of mouse knockouts. Moore B., et al. Communications Biology. Dec 21,2018. 1, 236.
 Many Newly Discovered Metabolism Genes Found in Mice
Many Newly Discovered Metabolism Genes Found in Mice
 Metabolism is the process of turning the food you eat into energy your body can use. Diseases that affect metabolism, including diabetes, are serious health concerns. While lifestyle choices and the environment play a role in how your metabolism operates, how many and which genes are involved in metabolism remain poorly understood. Using high-throughput mouse phenotyping, knockout mice – each lacking both copies of a specific gene – were studied for metabolic dysfunction by researchers from the Knockout Mouse Phenotyping Program (KOMP2), as part of the International Mouse Phenotyping Consortium (IMPC). With this method, they can test a single gene at a time to try to establish whether the missing gene is important to metabolism or other biological processes. Measuring things like blood glucose level, they found that 974 genes, out of 2016 tested, each had strong effects on metabolism. More importantly, almost half of those genes were newly discovered to be involved in metabolism. Furthermore, 51 of those genes had no known function until now.
Metabolism is the process of turning the food you eat into energy your body can use. Diseases that affect metabolism, including diabetes, are serious health concerns. While lifestyle choices and the environment play a role in how your metabolism operates, how many and which genes are involved in metabolism remain poorly understood. Using high-throughput mouse phenotyping, knockout mice – each lacking both copies of a specific gene – were studied for metabolic dysfunction by researchers from the Knockout Mouse Phenotyping Program (KOMP2), as part of the International Mouse Phenotyping Consortium (IMPC). With this method, they can test a single gene at a time to try to establish whether the missing gene is important to metabolism or other biological processes. Measuring things like blood glucose level, they found that 974 genes, out of 2016 tested, each had strong effects on metabolism. More importantly, almost half of those genes were newly discovered to be involved in metabolism. Furthermore, 51 of those genes had no known function until now.
This information may help researchers understand more about metabolic diseases, like diabetes, and develop better treatments. The genes identified in this study could be used, for example, as biomarkers for early diagnosis or personalized approaches for the treatment of metabolic diseases. Since this investigation only analyzed approximately 10% of all known genes, researchers will continue screening for genes associated with metabolic diseases in the coming years.
Reference:
 Mice Knockouts Provide Clues to Bad Breath and Beyond
Mice Knockouts Provide Clues to Bad Breath and Beyond
 A new study has helped to uncover a gene involved in serious cases of bad breath. Though halitosis (commonly known as bad breath) is most often caused by bacteria that colonize the mouth, rarer and more serious cases have no known cause. While halitosis can be a social annoyance, rarer forms can be the result of more concerning conditions, like liver cirrhosis. An international collaboration, including UC Davis of the Knockout Mouse Phenotyping Program (KOMP2), studied both mice and humans, revealing that mutations in gene called SELENBP1, encoding a novel human enzyme called methanethiol oxidase, can be the cause of this serious form of bad breath.
A new study has helped to uncover a gene involved in serious cases of bad breath. Though halitosis (commonly known as bad breath) is most often caused by bacteria that colonize the mouth, rarer and more serious cases have no known cause. While halitosis can be a social annoyance, rarer forms can be the result of more concerning conditions, like liver cirrhosis. An international collaboration, including UC Davis of the Knockout Mouse Phenotyping Program (KOMP2), studied both mice and humans, revealing that mutations in gene called SELENBP1, encoding a novel human enzyme called methanethiol oxidase, can be the cause of this serious form of bad breath.
Clues from patients indicated that sulfur metabolism genes likely play a role in the development of serious types of bad breath. And many of the patients had mutations in the gene SELENBP1. While this gene has been associated with some cancers, it has no known role in sulfur metabolism. Researchers then edited genes in mice to “knockout” the SELENBP1 gene. And though they didn’t directly smell the breath of the mice, they measured levels of sulfur containing compounds in the blood and urine. They found these levels increased compared to normal mice. They also confirmed biochemically that SELENBP1 does function as a sulfur metabolism enzyme and influences the concentrations of the biologically active molecules known to lead to bad breath. Given SELENBP1 association with cancer, this may also provide cancer researchers with new clues and targets for future study.
Reference:
Mutations in SELENBP1, encoding a novel human methanethiol oxidase, cause extraoral halitosis. Pol, A., Renkema, et al. Nature genetics. 2018 Jan;50(1):120-129.
 New Genes to Explore in Fight Against Hearing Loss
New Genes to Explore in Fight Against Hearing Loss
 More than 360 million people have some form of hearing impairment. We know that half of these cases are due to genetics, but the vast majority of genes responsible for many hearing loss syndromes are unknown. Now, research from the Knockout Mouse Phenotyping Program (KOMP2), part of the International Mouse Phenotyping Consortium (IMPC), shows that many more genes are involved in deafness than previously known. Because mice and humans share most genes, findings from mouse genes may tell us a lot about human hearing loss. Researchers tested over 3006 genetically modified mutant mice for hearing problems, exploring nearly 15% of the mouse genome. Hearing thresholds of the mice were measured with rising volumes of sound at different frequencies. While the study detected many genes already known to be involved in hearing loss, 52 of the genes were newly associated with hearing loss. These genes coded for many different types of proteins, from structural proteins to transcription factors, reflecting the complexity of the auditory system. Also, 41 of the 52 these genes were not even part of previously known networks of genes affecting hearing – highlighting potentially unexplored pathways of hearing loss. This information provides a large and unexplored genetic landscape that may help researchers understand more about hearing loss and develop better treatments.
More than 360 million people have some form of hearing impairment. We know that half of these cases are due to genetics, but the vast majority of genes responsible for many hearing loss syndromes are unknown. Now, research from the Knockout Mouse Phenotyping Program (KOMP2), part of the International Mouse Phenotyping Consortium (IMPC), shows that many more genes are involved in deafness than previously known. Because mice and humans share most genes, findings from mouse genes may tell us a lot about human hearing loss. Researchers tested over 3006 genetically modified mutant mice for hearing problems, exploring nearly 15% of the mouse genome. Hearing thresholds of the mice were measured with rising volumes of sound at different frequencies. While the study detected many genes already known to be involved in hearing loss, 52 of the genes were newly associated with hearing loss. These genes coded for many different types of proteins, from structural proteins to transcription factors, reflecting the complexity of the auditory system. Also, 41 of the 52 these genes were not even part of previously known networks of genes affecting hearing – highlighting potentially unexplored pathways of hearing loss. This information provides a large and unexplored genetic landscape that may help researchers understand more about hearing loss and develop better treatments.
Reference:
A large scale hearing loss screen reveals an extensive unexplored genetic landscape for auditory dysfunction. Bowl MR, Simon MM, Ingham NJ, Greenaway S, Santos L, Cater H, Taylor S, Mason J, Kurbatova N, Pearson S, Bower LR, Clary DA, Meziane H, Reilly P, Minowa O, Kelsey L, Tocchini-Valentini GP, Gao X, Bradley A, Skarnes WC, Moore M, Beaudet AL, Justice MJ, Seavitt J, Dickinson ME, Wurst W, de Angelis MH, Herault Y, Wakana S, Nutter LMJ, Flenniken AM, McKerlie C, Murray SA, Svenson KL, Braun RE, West DB, Lloyd KCK, Adams DJ, White J, Karp N, Flicek P, Smedley D, Meehan TF, Parkinson HE, Teboul LM, Wells S, Steel KP, Mallon AM, Brown SDM. Nature Communications. 2017 Oct 12;8 (886).
 Mouse Study Reveals Important Differences Between Males and Females
Mouse Study Reveals Important Differences Between Males and Females
 Historically, most researchers didn’t often study both sexes in their experiments, they assumed that results from male animals would be the same as female animals. However, we now know that sex influences the frequency, progression, and severity of the majority of common diseases and disorders, including cardiovascular and autoimmune diseases. Because of this, the NIH has mandated exploring sex as a biological variable, meaning researchers must consider sex as a biological variable in the design and analysis of their animal studies.
Historically, most researchers didn’t often study both sexes in their experiments, they assumed that results from male animals would be the same as female animals. However, we now know that sex influences the frequency, progression, and severity of the majority of common diseases and disorders, including cardiovascular and autoimmune diseases. Because of this, the NIH has mandated exploring sex as a biological variable, meaning researchers must consider sex as a biological variable in the design and analysis of their animal studies.
As part of the International Mouse Phenotyping Consortium (IMPC), Knockout Mouse Phenotyping Program (KOMP2) researchers supported by the Common Fund have explored how physical characteristics, vary by sex in normal and genetically modified mice. In the largest study of its kind, they analyzed 234 different physical characteristics in more than 50,000 mice. They found that the sex of the mice influenced many traits. For example, after accounting for weight, a known sexually dimorphic variable, 9.9% of categorical traits (things that can be put into categories, such as glucose tolerance) exhibited sexual dimorphism in normal mice. For continuous traits (things that can be measured on a scale, such as cholesterol levels), a far higher proportion exhibited sexual dimorphism at 56.6%. While some traits were expected to show differences in males and females, such as glucose levels and cardiac phenotypes, others were surprising and could not have been predicted. For example, vision abnormalities from the cornea were surprisingly found more often in female mice than males.
Not only did they study normal mice, but they also measured sexual dimorphism in many different genetically modified mice. To do this, they ”knocked out” different genes and measured whether any differences in the resulting physical traits depended on the sex. Unsurprisingly, some mutations only had effects in female mice, or vice versa. For example, only males and not females with the Usp47 gene knocked out, had high cholesterol levels, which would be important to consider in studies of heart disease or other diseases in which cholesterol is involved. The results have implications for the design of future animal studies which underpin research into treatments for human diseases. This study is a major step in highlighting the impact of sex differences in biomedicine and will help in accounting for those differences in the future biomedical studies.
Reference:
Prevalence of sexual dimorphism in mammalian phenotypic traits. Karp NA, Mason J, Beaudet AL, Benjamini Y, Bower L, Braun R E, Brown S DM, Chesler EJ, Dickinson ME, Flenniken AM, Fuchs H, de Angelis MH, Gao X, Guo S, Greenaway S, Heller R, Herault Y, Justice MJ, Kurbatova N, Lelliott CJ, Lloyd KC, Mallon A, Mank JE, Masuya H, McKerlie, TF Meehan, RF Mott, SA Murray, H Parkinson, R Ramirez-Solis, Santos, JR Seavitt, D Smedley C, Sorg T, Speak A O, Steel KP, Svenson L, The International Mouse Phenotyping Consortium, Wakana S, West D, Wells S, Westerberg H, Yaacoby S, White JK . Nature Communications. 2017 June 26;8 (15475).
 Researchers Use KOMP2 Mice to Learn About Genes That Help Repair Heart Tissue
Researchers Use KOMP2 Mice to Learn About Genes That Help Repair Heart Tissue
 The mammalian heart has the capacity to “self-renew,” or repair itself, only in very early life. Understanding more about the genes and mechanisms involved in heart repair during early life may provide clues into possible therapeutic approaches to reactivating this capacity in the adult heart. In a recent publication, Dr. James Martin and colleagues use mice generated from the Knockout Mouse Phenotyping Program (KOMP2) to study heart damage and repair. They identified a mechanism that promotes heart repair early in development through control of a gene called Pitx2. When the Pitx2 gene is activated, or “turned on,” it causes a cascade of other genes to also be activated, which limits damage and helps promote repair to the heart after an event like a heart attack. Ultimately, these researchers were able to increase expression of and activate Pitx2 in adult mice and promote some repair of heart tissue damage. These findings suggest possible mechanisms and genes to target when designing therapies to repair damaged heart tissue in humans.
The mammalian heart has the capacity to “self-renew,” or repair itself, only in very early life. Understanding more about the genes and mechanisms involved in heart repair during early life may provide clues into possible therapeutic approaches to reactivating this capacity in the adult heart. In a recent publication, Dr. James Martin and colleagues use mice generated from the Knockout Mouse Phenotyping Program (KOMP2) to study heart damage and repair. They identified a mechanism that promotes heart repair early in development through control of a gene called Pitx2. When the Pitx2 gene is activated, or “turned on,” it causes a cascade of other genes to also be activated, which limits damage and helps promote repair to the heart after an event like a heart attack. Ultimately, these researchers were able to increase expression of and activate Pitx2 in adult mice and promote some repair of heart tissue damage. These findings suggest possible mechanisms and genes to target when designing therapies to repair damaged heart tissue in humans.
Reference:
Pitx2 promotes heart repair by activating the antioxidant response after cardiac injury. Tao G, Kahr PC, Morikawa Y, Zhang M, Rahmani M, Heallen TR, Li L, Sun Z, Olson EN, Amendt BA, Martin JF. Nature. 2016 May 25;534(7605):119-23.
 Illuminating Essential Gene Function in Mice
Illuminating Essential Gene Function in Mice
 Drs. Mary Dickinson and Steve Murray and collaborators, as part of the Common Fund’s Knockout Mouse Phenotyping Program (KOMP2) and International Mouse Phenotyping Consortium (IMPC), have been using mouse models to study essential genes, or genes that are necessary for survival. In the absence of these genes, mice die as embryos, bringing up unique challenges for their analysis. However, this embryonic death also provides evidence for the critical role these essential genes play in normal growth and development in both mice and potentially humans. As part of the overall KOMP2/IMPC effort, researchers developed a strategy to more carefully study the function of these essential genes. In their strategy, they identify and describe the exact time of mouse lethality, assign phenotypes, and use a “reporter gene” to mimic the normal expression of the gene of interest in order to describe the function of these genes. This paper, High-throughput discovery of novel developmental phenotypes, is the first international, systematic effort to comprehensively characterize the functions of these genes in mice. This effort also includes 3D high-resolution imaging of embryos that are accessible on the IMPC portal. This imaging includes use of new and high level imaging that provides a level of detail not seen until now. For example, not much is currently known about disease associations with the gene Chtop. However, imaging of Chtop knockout embryos indicate abnormal eye development and neural tube defects, suggesting critical roles for this gene in normal eye and neural tube development. These results open a new line of research for scientists interested in studying development and diseases of the eye and nervous system. These new embryonic data add to the growing understanding of genetic mechanisms required for normal embryonic growth and development, while also providing insight into human developmental disorders and gene discovery for non-lethal conditions. As part of the full IMPC data, these data are contributing to this active and growing IMPC resource that will benefit the scientific community for years to come. Data are provided in real time to the entire research community, creating an “open access” environment allowing investigators to rapidly use the data to help their own research.
Drs. Mary Dickinson and Steve Murray and collaborators, as part of the Common Fund’s Knockout Mouse Phenotyping Program (KOMP2) and International Mouse Phenotyping Consortium (IMPC), have been using mouse models to study essential genes, or genes that are necessary for survival. In the absence of these genes, mice die as embryos, bringing up unique challenges for their analysis. However, this embryonic death also provides evidence for the critical role these essential genes play in normal growth and development in both mice and potentially humans. As part of the overall KOMP2/IMPC effort, researchers developed a strategy to more carefully study the function of these essential genes. In their strategy, they identify and describe the exact time of mouse lethality, assign phenotypes, and use a “reporter gene” to mimic the normal expression of the gene of interest in order to describe the function of these genes. This paper, High-throughput discovery of novel developmental phenotypes, is the first international, systematic effort to comprehensively characterize the functions of these genes in mice. This effort also includes 3D high-resolution imaging of embryos that are accessible on the IMPC portal. This imaging includes use of new and high level imaging that provides a level of detail not seen until now. For example, not much is currently known about disease associations with the gene Chtop. However, imaging of Chtop knockout embryos indicate abnormal eye development and neural tube defects, suggesting critical roles for this gene in normal eye and neural tube development. These results open a new line of research for scientists interested in studying development and diseases of the eye and nervous system. These new embryonic data add to the growing understanding of genetic mechanisms required for normal embryonic growth and development, while also providing insight into human developmental disorders and gene discovery for non-lethal conditions. As part of the full IMPC data, these data are contributing to this active and growing IMPC resource that will benefit the scientific community for years to come. Data are provided in real time to the entire research community, creating an “open access” environment allowing investigators to rapidly use the data to help their own research.
More information about the mice phenotyped through IMPC here.
Reference:
High-throughput discovery of novel developmental phenotypes. Dickinson ME, Ann M, Flenniken AM, Ji ,et al. Nature. 2016 Sep 14;537(7621):508-514.
 IMPC Leads Coordinated Effort to Characterize Every Gene in the Mouse Genome
IMPC Leads Coordinated Effort to Characterize Every Gene in the Mouse Genome
 The International Mouse Phenotyping Consortium (IMPC) was featured in Biocentury Innovations. The article highlighted its history, organization, and exciting progress made to date. The IMPC is a confederation of international mouse phenotyping projects, including the NIH Common Fund’s Knockout Mouse Phenotyping Program (KOMP2). The goal of the IMPC is to generate knockout lines for the 21,000 known and predicted mouse genes, to ascribe functions to as many of those genes as possible through systematic phenotyping, and to distribute the data freely to the scientific community. Once complete, this will be a critical resource to researchers studying mice as model organisms and will help ascribe functions to related human genes that are currently uncharacterized. This report highlights some of the major advantages of broad-based, systemic phenotyping, including standardization of phenotyping assays and the ability to detect phenotypes in a variety of tissues for the same gene. The work of the IMPC may also inform large scale efforts to generate, coordinate, and analyze human data, such as the Precision Medicine Initiative.
The International Mouse Phenotyping Consortium (IMPC) was featured in Biocentury Innovations. The article highlighted its history, organization, and exciting progress made to date. The IMPC is a confederation of international mouse phenotyping projects, including the NIH Common Fund’s Knockout Mouse Phenotyping Program (KOMP2). The goal of the IMPC is to generate knockout lines for the 21,000 known and predicted mouse genes, to ascribe functions to as many of those genes as possible through systematic phenotyping, and to distribute the data freely to the scientific community. Once complete, this will be a critical resource to researchers studying mice as model organisms and will help ascribe functions to related human genes that are currently uncharacterized. This report highlights some of the major advantages of broad-based, systemic phenotyping, including standardization of phenotyping assays and the ability to detect phenotypes in a variety of tissues for the same gene. The work of the IMPC may also inform large scale efforts to generate, coordinate, and analyze human data, such as the Precision Medicine Initiative.
More information about the mice phenotyped through IMPC here.
Reference:
Knocks Heard ‘Round The World. Tkach, K. Biocentury Innovations. 2015 September 24.
 New Statistical Tools for Standardizing High Throughput Analysis in Mouse Phenotyping
New Statistical Tools for Standardizing High Throughput Analysis in Mouse Phenotyping

Analyzing large amounts of data can be a daunting task in any research field; this also includes applying appropriate statistical analyses to ensure the most robust conclusions can be drawn. With many areas of research now generating massive data sets, there is a growing need for accessible and freely available statistical tools. One tool recently developed by Kurbatova et al., PhenStat, is a software statistical software package designed specifically for phenotyping data. The software uses the freely available R package to provide a variety of statistical methods for the identification of phenotypic associations. R is a widely available platform commonly used in bioinformatics.
The PhenStat software itself is specifically designed for the type of high throughput data generated by large research programs, but is also suitable for small and large scale users. The software package has been tested and implemented by the KOMP2 program as part of the International Mouse Phenotyping Consortium (IMPC), which aims to produce knockout mice and carry out high-throughput phenotyping of each line in order to determine the function of every gene in the mouse genome. The consortium and its researchers are increasingly focused on reproducibility, studying both sexes, and using appropriate statistical tools and methodologies. This software aims to provide the best tools to assist in these important efforts by helping researchers produce the most robust analysis. With appropriate and well documented analysis, an increase in reproducibility of results by other parties can be enhanced, which is important for all research, but particularly for in-vivo studies where limiting expensive, redundant, and unnecessary experimentation is essential.
Reference:
PhenStat: A Tool Kit for Standardized Analysis of High Throughput Phenotypic Data. Kurbatova, N., Mason, J. C., Morgan, H., Meehan, T. F., & Karp, N. A. PloS one. 2014 Jul 6;.10(7), e0131274-e0131274.
 NIH-Funded KOMP2 Program Leads the Way in Efforts to Ensure Reproducibility and Enhance Transparency in Research
NIH-Funded KOMP2 Program Leads the Way in Efforts to Ensure Reproducibility and Enhance Transparency in Research
Researchers funded by the National Institutes of Health’s Common Fund Knockout Mouse Phenotyping Program (KOMP2) are making efforts to improve methods of data collection and reporting in animal research, with the goals of making these processes more accessible and research findings more reproducible. The KOMP2 program, a part of the International Mouse Phenotyping Consortium (IMPC), aims to extensively test and generate data about mice with disrupted, or “knocked out,” genes to help better understand human biology and disease. KOMP2 researchers are examining the clinical and physical characteristics – phenotypes – of mice carrying mutations in approximately 20,000 specific genes across the genome. They hope to systematically describe each gene and its biological function.
In 2010, the United Kingdom National Centre for the Replacement, Refinement, and Reduction of Animals in Research (3R) introduced the ARRIVE (Animal Research: Reporting of In Vivo Experiments) guidelines ![]() . These guidelines were designed to address an emerging problem in biomedical animal studies -- a lack of reproducibility of research results -- and improve the overall communication of research findings. The ARRIVE guidelines consist of a checklist to be used when submitting manuscripts that include animal research. They lay out reporting requirements to ensure all the information is available to allow fully reproducible research.
. These guidelines were designed to address an emerging problem in biomedical animal studies -- a lack of reproducibility of research results -- and improve the overall communication of research findings. The ARRIVE guidelines consist of a checklist to be used when submitting manuscripts that include animal research. They lay out reporting requirements to ensure all the information is available to allow fully reproducible research.
To enhance and embrace reproducibility in the IMPC large-scale research data collection and analyses efforts, KOMP2-funded investigators modified the ARRIVE guidelines to apply them to the large international database housing all the program data ![]() . The investigators described the process and challenges of applying the guidelines to the IMPC database May 20, 2015, in PLoS Biology. Their efforts assessed and documented how each of the IMPC centers carried out experimental procedures; details on how the data were obtained are available for download from the IMPC web portal. Investigators also developed a new and standardized language for the IMPC consortium, allowing all phenotypic data to be described in the same way and compared across research sites.
. The investigators described the process and challenges of applying the guidelines to the IMPC database May 20, 2015, in PLoS Biology. Their efforts assessed and documented how each of the IMPC centers carried out experimental procedures; details on how the data were obtained are available for download from the IMPC web portal. Investigators also developed a new and standardized language for the IMPC consortium, allowing all phenotypic data to be described in the same way and compared across research sites.
Reference:
Applying the ARRIVE Guidelines to an In Vivo Database. Karp N, Meehan T, Morgan H, Mason J, Blake A, Kurbatova N, Smedley D, Jacobsen J, Mott R, Iyer V, Matthews P, Melvin D, Wells S, Flenniken A, Masuya H, Wakana S, White J, Lloyd KC, Reynolds C, Paylor R, West D, Svenson K, Chesler E, de Angelis M, Tocchini-Valentini G, Sorg T, Herault Y, Parkinson H, Mallon A, Brown S. PLOS Biology. 2015 May 20.
 A Comprehensive New Atlas for Knockout Mice Phenotypes
A Comprehensive New Atlas for Knockout Mice Phenotypes
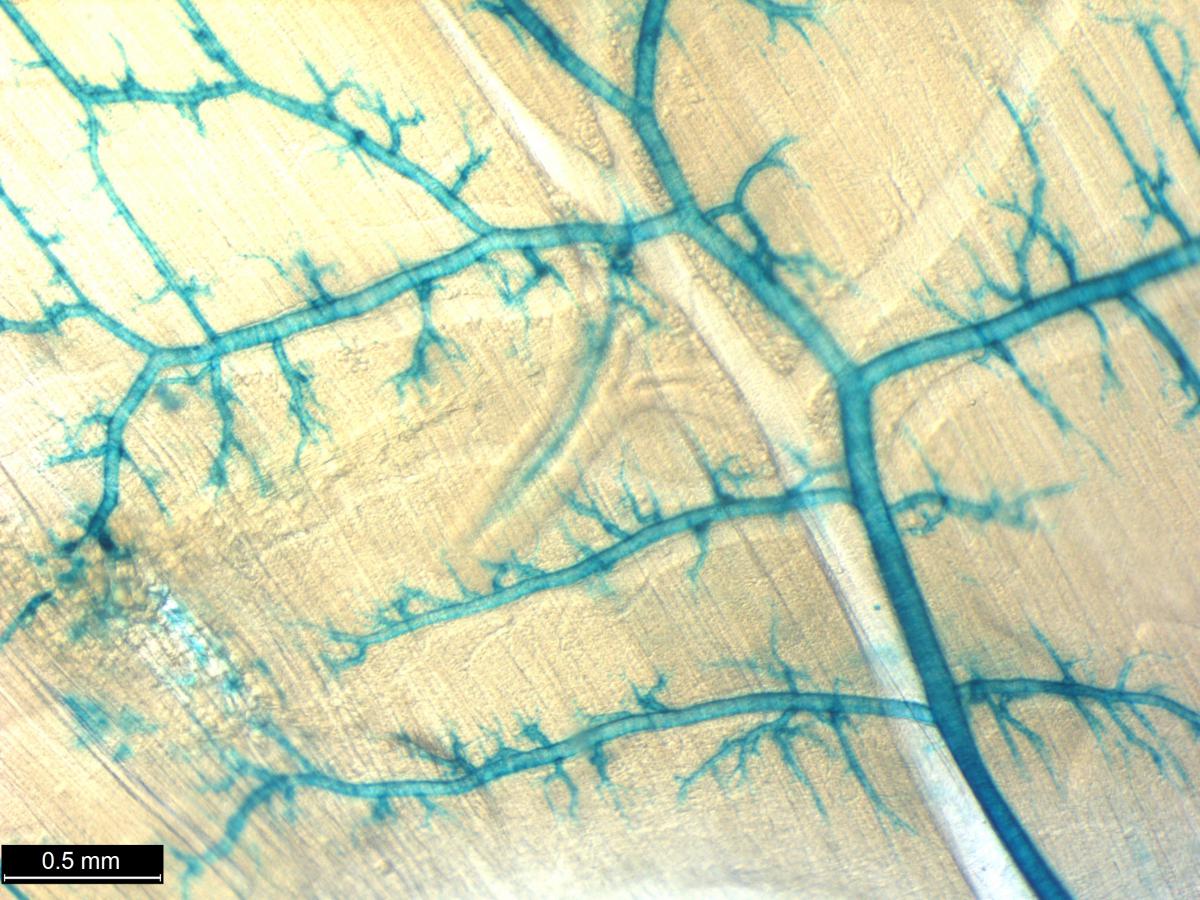
Researchers from the International Mouse Phenotyping Consortium (IPMC), an international effort that includes NIH funded researchers from the Common Fund Knockout Mouse Phenotyping Program (KOMP2) have developed an atlas detailing gene expression patterns in 313 different mutant mouse lines. The Consortium as a whole is currently on track to develop 8,500 mutant mice and this manuscript represents one of the first comprehensive glimpses at early expression data coming from this group of investigators. Researchers generated the mutant mouse lines using embryonic stem cells (ESCs) in which the gene of interested is replaced with a so called “reporter gene” in such a way that the reporter mimics the normal expression of the gene of interest. A simple chemical reaction on the samples results in a blue stain where the gene is expressed. This well-known LacZ reporter system can then be used to compare various staining patterns in the mice. These staining strategies were carried out in both whole mount and on frozen sections of male and female mice and embryos. The robustness and repeatability of the lacZ staining in both 3D and in sections confirmed the feasibility of using this reporter to reveal the expression of many poorly annotated genes. The generated atlas includes information on the location and possible function of many genes that currently have unknown functions.
In this study, many large organs, such as the brain, were described in remarkable detail providing new and highly specialized information on the localization patterns of gene expression for many genes. Many organs also had complex and unique staining patterns, for example, not only was a large percentage of all reporter gene expression found in the testis, but 8% of all reporter expression was found restricted exclusively in the testis. One gene with unique expression in the testis was Jazf, which has been previously implicated in endometrial cancer, but has yet to have any known role in testicular function. Overall, the atlas provides an exceptional and comprehensive resource describing the anatomical location of gene expression in mice and provides insight into unique and unknown functions that can be targeted for further study. The full resource, to be completed by the IMPC, will provide even deeper levels of detail and data for the 8,500 knockout mouse lines that will complement this effort and give insight into the roles of thousands of genes in embryonic development, adult physiology, aging, and disease.
Reference:
A lacZ reporter gene expression atlas for 313 adult KOMP mutant mouse lines. West, D. B., R. K. Pasumarthi, B. Baridon, E. Djan, A. Trainor, S. M. Griffey, E. K. Engelhard, J. Rapp, B. Li, P. J. Jong and K. C. Lloyd. Genome Res. 2015;25(4): 598-607. PMID: 25591789.
 Applying a New Method to Generate Knockout Mice
Applying a New Method to Generate Knockout Mice
Research supported by the Common Fund’s Knockout Mouse Phenotyping Program (KOMP2) has applied a  new way to generate conditional knockout mice, which are mice with a gene that can be selectively eliminated in specific cells or tissues at a specific point in time. Led by Dr. K.C. Kent Lloyd, researchers at the University of California Davis modified a technology known as CRISPR, or Clustered Regularly Interspaced Short Palindromic Repeat, that uses components of a bacterial immune system to precisely target genes for deletion (for more information on CRISPR, see this New York Times article). In this new study, scientists modified the CRISPR system to enhance the specificity of gene targeting while also introducing genetic changes that allow the target gene to be conditionally deleted (or “knocked out”), rather than knocked out in all cells from the beginning of development. Researchers used the new technique to generate a conditional knockout of the Isoprenoid synthase containing domain (Ispd) gene. Mutations in the human ISPD gene are associated with the neurodevelopmental disorder Walker-Warburg syndrome, a severe form of congenital muscular dystrophy affecting the muscles, brain, and eyes, that typically results in death by 3 years of age. Ispd knockout mice die at birth, limiting their effectiveness in scientific research. The conditional Ispd knockout mice may allow scientists to distinguish the effects of Ispd deletion in disease-relevant tissues at different stages of development. Beyond this important gene, however, the new technique may allow easier, faster, and more efficient generation of conditional knockout mice for many genes, both within the KOMP2 project and in labs across the world.
new way to generate conditional knockout mice, which are mice with a gene that can be selectively eliminated in specific cells or tissues at a specific point in time. Led by Dr. K.C. Kent Lloyd, researchers at the University of California Davis modified a technology known as CRISPR, or Clustered Regularly Interspaced Short Palindromic Repeat, that uses components of a bacterial immune system to precisely target genes for deletion (for more information on CRISPR, see this New York Times article). In this new study, scientists modified the CRISPR system to enhance the specificity of gene targeting while also introducing genetic changes that allow the target gene to be conditionally deleted (or “knocked out”), rather than knocked out in all cells from the beginning of development. Researchers used the new technique to generate a conditional knockout of the Isoprenoid synthase containing domain (Ispd) gene. Mutations in the human ISPD gene are associated with the neurodevelopmental disorder Walker-Warburg syndrome, a severe form of congenital muscular dystrophy affecting the muscles, brain, and eyes, that typically results in death by 3 years of age. Ispd knockout mice die at birth, limiting their effectiveness in scientific research. The conditional Ispd knockout mice may allow scientists to distinguish the effects of Ispd deletion in disease-relevant tissues at different stages of development. Beyond this important gene, however, the new technique may allow easier, faster, and more efficient generation of conditional knockout mice for many genes, both within the KOMP2 project and in labs across the world.
Reference:
Conditional targeting of Ispd using paired Cas9 nickase and a single DNA template in mice. FEBS Open Biology. Lee, AY and Lloyd, KC. July 2014; 4: 637-42. PMID: 25161872.
 “Perfect Host” Developed!
“Perfect Host” Developed!
Investigators at The Jackson Laboratory have developed a novel approach that enables highly efficient generation of mouse strains from Embryonic Stem (ES) cells. This advance overcomes a major limitation in mutant mouse production; namely, the uncertainty of whether the desired mutation created in ES cells is incorporated into the mouse’s reproductive cells so that it is passed along to offspring. This process has been slow, laborious, and expensive. The Perfect Host technology ensures that chimeras made by injecting ES cells into host embryos have gametes derived only from the ES cells carrying the desired mutation. Thus all offspring are paternally derived from the ES cells and will carry the mutation. This research was made possible through support from The Jackson Laboratory, the Maine Technology Institute, in addition to several NIH Institutes and Centers, including KOMP and the use of KOMP-generated ESC cell lines. This new technology will be extremely useful for the KOMP2 program, by reducing the number of mice needed for each experiment, and greatly increasing the speed and efficiency of mouse production.
 Interview with KOMP2 awardee Dr. Monica Justice
Interview with KOMP2 awardee Dr. Monica Justice
Read an interview with KOMP2 awardee Dr. Monica Justice from Baylor College of Medicine, in which she discusses her career path, current work in hematopoietic cancers and genetic syndromes, and future goals for the KOMP2 project. Find out how her early years on the farm inspired a love of biology, what experiences sparked her interest in pediatric medicine, and how she enjoys spending time outside of the lab.
Read Of Mice and Men, and Medicine: an interview with Monica Justice.
 Funding Opportunity Announcement to phenotype embryonic lethal knockout mice from the International Mouse Phenotyping Consortium (IMPC)!
Funding Opportunity Announcement to phenotype embryonic lethal knockout mice from the International Mouse Phenotyping Consortium (IMPC)!
The IMPC, a worldwide consortium that includes KOMP2 as a member, is generating an estimated 20,000 knockout mouse strains, up to 30% of which are expected to be embryonic or perinatal lethal. This funding opportunity announcement invites applications to phenotype embryonic lethal mouse strains with a long term goal of revealing important insights into normal development in addition to a variety of diseases. This funding opportunity is supported by several NIH Institutes and Centers, and is related to, but separate from, the Common Fund’s KOMP2 program.
Read the Funding Opportunity here.
 Updates from KOMP2
Updates from KOMP2
See what KOMP2 has been doing lately! Download slides about KOMP2 history, current status, and future directions here.
 KOMP2 Program Makes First Awards
KOMP2 Program Makes First Awards
Seven new projects receive NIH Common Fund support in fiscal year 2011 via the KOMP2 program. The program is creating a valuable genetic resource for revealing mammalian gene function, providing insight into genes that affect human health and disease.
Read more…



