Science Highlights
NIGMS Image Gallery
NIH Common Fund awardee wins Nobel Prize in Chemistry
Dr. Brian Kobilka, a grantee of the Common Fund Structural Biology program, has been awarded the Nobel Prize in Chemistry along with Dr. Robert Lefkowitz for their groundbreaking studies of G-protein coupled receptors (GPCRs). GPCRs are proteins located in cell membranes that transmit important information about the environment outside the cell, including light, smell, taste, hormone levels, and neurotransmitters. GPCRs mediate a wide range of medically-relevant biological processes, and are the target of almost half of pharmaceuticals on the market (see also “Why care about GPCRs?”). A major goal of the Structural Biology program is to determine the unique three-dimensional structure of membrane proteins such as GPCRs, in order to inform the design of therapeutics that specifically fit inside the target protein, thereby improving drug efficacy and reducing side effects.
Dr. Kobilka’s Common Fund-supported research helped develop new methods for producing and stabilizing GPCRs in large enough amounts to allow scientists to precisely determine their protein structures. Dr. Kobilka and colleagues at Stanford University pioneered the use of a novel approach to protein structure determination that involved fusing a small protein called T4 lysozyme, or T4L, into a part of GPCRs that is very flexible and therefore poses challenges to structure determination. He first used this technique in collaboration with Dr. Raymond Stevens and colleagues at The Scripps Research Institute to determine the structure of the β2-adrenergic receptor (β2AR), a protein critical in cardiac and pulmonary function. The T4L fusion strategy has proven to be broadly applicable in determining the structure of a number of biologically important GPCRs involved in a wide range of diseases, including many structures discovered by researchers in the Common Fund Structural Biology program. Dr. Kobilka’s catalytic and transformative approach to protein structure determination is an excellent example of the nature of Common Fund supported research, which aims to accelerate biomedical research that is relevant for multiple diseases and conditions. In addition to support from the Common Fund Structural Biology program, Dr. Kobilka has also received support from the National Institute of General Medical Sciences, the National Institute of Neurological Disorders and Stroke, and the National Heart, Lung, and Blood Institute. Dr. Lefkowitz has received support from the National Heart, Lung, and Blood Institute as well.
June 7, 2012 
NEW! Structural Biology researchers publish methods and tools to aid in protein structure determination!
The Structural Biology Program has published a special METHODS (link is external) issue on “Membrane Protein Technologies for Structural Biology.” This issue contains 18 methods papers that originated from the 2010 Structural Biology investigators meeting, where scientific approaches and research results were shared among meeting attendees.
Reference:
Membrane Protein Technologies for Structural Biology. Methods, December 2011; Volume 55, Issue 4, Pages 271-420.
Dr. Raymond Stevens and colleagues have published a “fusion partner (link is external)toolchest (link is external)” to aid in the stabilization and crystallization of G protein-coupled receptors. This paper provides data on five selected fusion partners, including one (BRIL) that is a great improvement over the previously utilized T4 lysozyme fusion partner.
Reference:
Chun E, Thompson AA, Liu W, Roth CB, Griffith MT, Katritch V, Kunken J, Cherezov V, Hanson MA, Stevens RC. Structure, 2012 June 6; 20(6): 967-976.
May 31, 2012 
RESEARCHERS DISCOVER STRUCTURE OF OPIOID RECEPTORS
Opioid receptors are proteins found on the surface of cells in the nervous and digestive systems that bind opioid proteins, molecules that naturally occur in the body and play a role in regulating pain, pleasure, mood, addiction, and digestion. An array of legal and illegal drugs such as morphine, codeine, and heroin also bind to these receptors and control their activity, but usually with unwanted side effects such as hallucinations and addiction, which limits their clinical use. The development of selective therapeutics that control the activity of opioid receptors without these side effects holds great promise as pain relievers, anti-depressants, and anti-anxiety treatments. The development of such agents could have a revolutionizing effect on the treatment of acute and chronic pain, several neuropsychiatric disorders, and addiction.
Dr. Raymond Stevens, partly funded by the NIH Common Fund’s Structural Biology program and the National Institute of General Medical Science’s Protein Structure Initiative, along with colleagues, has published the three-dimensional structures of two members of the human opioid receptor family- the kappa opioid receptor (KOR) and the nociceptin/orphanin FQ peptide receptor (NOP). As reported in the May 17, 2012 issue of Nature, these structures reveal unprecedented detail about the shape of these receptors, which may allow researchers to design drugs that can interact with these receptors in specific ways to elicit only the desired effects. KOR is the only receptor that binds the active ingredient in the plant Salvia divinorum (also known as “Salvia” or “Magic Mint”), which has recently gained popularity as a recreational drug of abuse causing hallucinations and psychedelic experiences (see the National Institute of Drug Abuse InfoFacts: Salvia). The part of the receptors where drugs and other molecules bind, called the binding pocket, is very large in both KOR and NOP. KOR and NOP differ in only a few specific places within the binding pocket, but these differences result in significant changes in the shape of the pocket, explaining why some molecules specifically bind to one receptor, but not the other. In the same issue of Nature, Dr. Brian Kobilka and colleagues published the structures of the mu and delta opioid receptors; collectively, these four papers reveal the structures of the entire family of human opioid receptors. Drs. Stevens and Kobilka used sophisticated techniques, developed in part through previous Common Fund support, to create the protein crystals needed to reveal the underlying protein structure. These studies provide a major clue in understanding the selectivity of opioid receptors, opening up new avenues of research into basic research about brain function and consciousness, as well as the development of clinically useful therapeutics.
References:
- Wu H, Wacker D, Katritch V, Mileni M, Han GW, Vardy E, Liu W, Thompson AA, Huang XP, Carroll FI, Mascarella SW, Westkaemper RB, Mosier PD, Roth BL, Cherezov V, Stevens RC. Structure of the human kappa opioid receptor in complex with JDTic. Nature, 2012 Mar 21 (online publication date); 485(7398): 327-32. PMID: 22437504.
- Thompson AA, Liu W, Chun E, Katritch V, Wu H, Vardy E, Huang X-P, Trapella C, Guerrini R, Calo G, Roth BL, Cherezov V, Stevens RC. Structure of the nociceptin/orphanin FQ receptor in complex with a peptide mimetic. Nature, 2012 May 16; 485(7398):395-9. PMID: 22596163.
- Manglik A, Kruse AC, Kobilka TS, Thian FS, Mathiesen JM, Sunahara RK, Pardo L, Weis WI, Kobilka BK, Granier S. Crystal structure of the mu-opioid receptor bound to a morphinan antagonist. Nature, 2012 Mar 21 (online publication date); 485(7398): 321-6. PMID: 22437502.
- Granier S, Manglik A, Kruse AC, Kobilka TS, Thian FS, Weis WI, Kobilka B. Structure of the delta-opioid receptor bound to naltrindole. Nature, 2012 May 16; 485(7398): 400-4. PMID: 22596164.
Common Fund Researchers Uncover Structure of Important Target for Drug Design

Researchers supported by the NIH Common Fund and the National Institute of General Medical Sciences teamed up to characterize an important G Protein-coupled receptor (GPCR). GPCRs are a class of membrane proteins involved in an array of physiological functions and human diseases, including multiple sclerosis. Importantly, these receptor proteins are the target of approximately 40% of all medications currently on the market. Using technology developed in part through support from the Common Fund’s Structural Biology Program, the research teams of Drs. Raymond Stevens and Hugh Rosen at The Scripps Research Institute teamed up to examine the GPCR sphingosine 1-phosphate 1 (S1P1) bound to different activators and inhibitors developed through the Common Fund’s Molecular Libraries and Imaging Program. S1P1 receptors play critical roles in controlling multiple sclerosis and other diseases, making the discovery useful for advancing treatment options. Advances made in this study allowed the researchers to identify how molecules bind in different ways to the receptor and better understand at a detailed level how access to the binding pocket is gained. These advances pave the way for more targeted drug design that will yield highly effective therapeutics with fewer side effects than current treatments. These findings were published in the February 17, 2012 issue of the journal Science.
- Read the NIH press release
- Read more about the Molecular Libraries and Imaging program…
- Read more about the Structural Biology program
References:
- Hanson MA, Roth CB, Jo E, Griffith MT, Scott FL, Reinhart G, Desale H, Clemons B, Cahalan SM, Schuerer SC, Sanna MG, Han GW, Kuhn P, Rosen H, Stevens RC. Crystal structure of lipid G protein-coupled receptor. Science, 2012 Feb 17; 335(6070): 851-5. PMID: 22344443.
Structure of allergy-causing histamine receptor revealed

As millions of allergy sufferers know, common substances like pollen, mold, and pet hair can trigger an over-reaction of the immune system that results in sneezing, congestion, and eye irritation. Dr. Raymond Stevens and colleagues at the Joint Center for Innovative Membrane Protein Technologies at the Scripps Research Institute, supported in part by the Structural Biology program, have determined the three-dimensional structure of the histamine receptor responsible for allergic reactions, H1R. This structure reveals important clues about how anti-histamine drugs interact with H1R to dampen allergic reactions, and also why some anti-histamine drugs have more side effects than others.
Described in the July 7, 2011 issue of the journal Nature, Dr. Stevens and colleagues used methods pioneered by the Structural Biology program to determine the structure of H1R interacting with the anti-histamine drug doxepin. Doxepin, one of the early anti-histamine drugs, effectively blocks an allergic reaction but also causes many side effects such as sedation, dry mouth, and heart beat irregularities. These side effects are partially due to doxepin’s interaction with other proteins besides H1R. Dr. Stevens’ research shows that doxepin interacts with a piece of H1R that is very similar in shape to pieces of other, closely related proteins. However, this study also revealed small differences in shape between H1R and closely related proteins, including a larger binding “pocket” in H1R for an anti-histamine drug to fit snugly inside. Dr. Stevens then used computer modeling to show that newer anti-histamine drugs exploit these differences to fit perfectly with H1R but not other receptors, explaining why they produce fewer side effects. A better understanding of protein structure can help scientists develop more effective drugs with fewer side effects, a major goal of the Structural Biology program.
Reference:
- Shimamura T, Shiroishi M, Weyand S, Tsujimoto H, Winter G, Katritch V, Abagyan R, Cherezov V, Liu W, Han GW, Kobayashi T, Stevens RC, and Iwata S. Structure of the human histamine H1 receptor complex with doxepin. Nature 2011. 475: 65-70. PMID: 21697825.
Researcher Uncovers Shape of Protein Important for Neurological Disorders

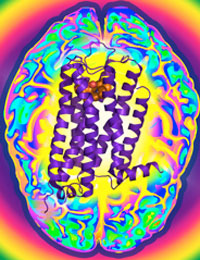
Dopamine is an essential "neurotransmitter", conveying important signals from one cell in the brain to another. Disruption of these signals initiated by dopamine can lead to schizophrenia, Parkinson’s disease, or drug addiction. The D3R protein is one of five dopamine receptor subtypes that bind dopamine to mediate cellular communication and is a primary target for the development of drugs to treat these conditions. Frequently, however, drugs that affect D3R can also affect the other dopamine receptor proteins and causing unwanted side effects. The Joint Center for Integral Membrane Protein Technologies-Complexes (JCIMPT-Complexes) at the Scripps Research Institute headed by Dr. Raymond C. Stevens, funded in part by the Common Fund’s Structural Biology program, has determined the three-dimensional shape of D3R, described in the November 19th advance online issue of the journal Science. This shape reveals subtle differences between D3R and the closely related protein D2R, which may assist researchers in designing effective D3R-specific drugs with fewer side effects. The research finding is particularly significant because D3R is a type of "membrane protein," a protein that is embedded in the viscous lipid environment surrounding a cell, making it challenging to isolate the protein to determine its shape. This finding comes soon after Dr. Stevens’ discovery of the shape of CXCR4, a protein important for HIV infection and cancer. By using methods similar to those pioneered by JCIMPT-Complexes and other research labs within the Structural Biology program, scientists have the potential to determine the structure of Image courtesy of Dr. Raymond Stevens many more proteins implicated in human diseases and develop more targeted therapeutic drugs.
Reference:
- Chien EY, Liu W, Zhao Q, Katritch V, Han GW, Hanson MA, Shi L, Newman AH, Javitch JA, Cherezov V, Stevens RC. Structure of the human dopamine D3 receptor in complex with a D2/D3 selective antagonist. Science 2010 Nov 19; 330(6007): 1091-5. PMID: 21097933.
Researcher Discovers Shape of Proteins Important in HIV and Cancer

 The Joint Center for Integral Membrane Protein Technologies-Complexes (JCIMPT-Complexes) at the Scripps Research Institute headed by Dr. Raymond C. Stevens, funded in part by the Common Fund’s Structural Biology Program, has determined the three-dimensional structure of CXCR4, a cellular protein important for HIV infection as well as the growth and metastasis of many types of cancer. A recent paper by Dr. Stevens and colleagues, published in the October 7th advance online issue of the journal Science, describes the molecular structure of the CXCR4 protein bound to molecules known to inhibit CXCR4’s function. This study reveals that the location and the shape of the sites where inhibitory molecules bind is very different between CXCR4 and other closely related proteins. Using knowledge of these binding sites, researchers may one day develop new strategies to design drugs that bind CXCR4 to block HIV infection or stall the spread of some cancers.
The Joint Center for Integral Membrane Protein Technologies-Complexes (JCIMPT-Complexes) at the Scripps Research Institute headed by Dr. Raymond C. Stevens, funded in part by the Common Fund’s Structural Biology Program, has determined the three-dimensional structure of CXCR4, a cellular protein important for HIV infection as well as the growth and metastasis of many types of cancer. A recent paper by Dr. Stevens and colleagues, published in the October 7th advance online issue of the journal Science, describes the molecular structure of the CXCR4 protein bound to molecules known to inhibit CXCR4’s function. This study reveals that the location and the shape of the sites where inhibitory molecules bind is very different between CXCR4 and other closely related proteins. Using knowledge of these binding sites, researchers may one day develop new strategies to design drugs that bind CXCR4 to block HIV infection or stall the spread of some cancers.
Reference:
Wu B, Chien EY, Mol CD, Fenalti G, Liu W, Katritch V, Abagyan R, Brooun A, Wells P, Bi FC, Hamel DJ, Kuhn P, Handel TM, Cherezov V, Stevens RC. Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists. Science 2010 Nov 19; 330(6007): 1066-71. PMID: 20929726.
Scientists Develop Breakthrough Method for Finding Protein Shapes
 An international team of researchers, including Dr. Petra Fromme of the Common Fund’s Structural Biology program, has reported a novel method for determining protein structure that overcomes many of the technical difficulties associated with previous methods. The new method, reported in the February 3, 2011 issue of Nature, uses smaller amounts of protein samples, causes less damage to the samples, and is much faster than traditional methods. Using this new method, Dr. Fromme and colleagues determined the structure of photosystem 1, a large and highly complex protein used by plants during photosynthesis to convert carbon dioxide into sugar using energy from the sun. This structure matched the previously reported structure of photosystem 1 obtained using traditional methods, demonstrating the accuracy of the new method. This new method should allow scientists to determine the structures of many different kinds of proteins, including those that have been resistant to traditional approaches. It is particularly difficult to determine the structure of proteins embedded within cell membranes, yet these proteins play critical roles in health and disease and are common drug targets. The Common Fund’s Structural Biology program seeks new ways to determine the structure of membrane proteins, in the hopes of using this information to design drugs that will bind to a well-defined structural feature, producing fewer side effects. The breakthrough method developed by Dr. Fromme and colleagues will likely be a powerful tool in determining the structure of numerous membrane proteins.
An international team of researchers, including Dr. Petra Fromme of the Common Fund’s Structural Biology program, has reported a novel method for determining protein structure that overcomes many of the technical difficulties associated with previous methods. The new method, reported in the February 3, 2011 issue of Nature, uses smaller amounts of protein samples, causes less damage to the samples, and is much faster than traditional methods. Using this new method, Dr. Fromme and colleagues determined the structure of photosystem 1, a large and highly complex protein used by plants during photosynthesis to convert carbon dioxide into sugar using energy from the sun. This structure matched the previously reported structure of photosystem 1 obtained using traditional methods, demonstrating the accuracy of the new method. This new method should allow scientists to determine the structures of many different kinds of proteins, including those that have been resistant to traditional approaches. It is particularly difficult to determine the structure of proteins embedded within cell membranes, yet these proteins play critical roles in health and disease and are common drug targets. The Common Fund’s Structural Biology program seeks new ways to determine the structure of membrane proteins, in the hopes of using this information to design drugs that will bind to a well-defined structural feature, producing fewer side effects. The breakthrough method developed by Dr. Fromme and colleagues will likely be a powerful tool in determining the structure of numerous membrane proteins.
Reference:
- Chapman HN, Fromme P, Barty A, White TA, Kirian RA, et al. Femtosecond X-ray protein nanocrystallography. Nature, 2011 Feb 3; 470(7332): 73-7. PMID: 21293373.
Tiny Device Helps View Protein Structure
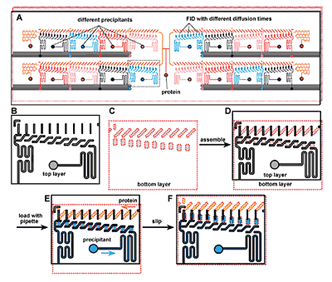
A "SlipChip"; designed to screen a protein against 16 different precipitants using the free interface diffusion (FID) method of crystallization. Reproduced with permission from the ACS.
Researchers in the Common Fund's Structural Biology Research program have engineered a tiny new device to optimize the conditions needed for proteins to form crystals so their function can be studied in cells. Knowing a protein's structure can provide clues about how it functions normally, how it behaves abnormally in a diseased cell, and how its function may be restored by drugs that target specific parts of the protein that control activity
X-ray crystallography is a powerful tool to make high resolution images of protein structure by identifying the position of individual atoms in three-dimensional (3-D) space and then converting them into a topological map of the protein. However, for X-ray crystallography to work, the protein molecules must be concentrated and stacked in an orderly fashion into a protein crystal. Otherwise, the protein molecules just flop around, making it impossible to accurately pinpoint the position of the individual atoms. This is challenging because proteins are very finicky about the conditions under which they will form well-ordered crystals and very large amounts of proteins are usually needed to form crystals for analysis of 3-D structure.
Dr. Rustem Ismagilov at the University of Chicago has developed a microfluidic device called a "SlipChip" that allows researchers to explore the ideal conditions that can support the crystallization of a protein using very small amounts. The SlipChip can simultaneously test 160 different conditions that may support protein crystal formation. The protein of interest is injected in to a central port on the SlipChip where it flows through microfluidic channels and enters 160 nanometer-scale sized wells grouped into 16 separate units containing 10 wells each (see figure). The protein in each unit is subsequently exposed to a different precipitant, that is, a chemical solution that may support crystallization. This occurs after each precipitant is injected into one of the 16 units where it flows into 10 wells that sit underneath the protein-containing wells and the two are brought in to contact by "slipping"; the protein wells closer to the precipitant wells. The physical distance between the protein and precipitant wells on the SlipChip gradually increases from the first well to the tenth well, varying the time the protein and precipitant interact with each other. Precise control of this timing is important for some proteins that will not form well-ordered crystals if the interaction time is too fast. The beauty of the SlipChip is that it increases the chances of finding the optimum conditions for proteins and precipitants to interact so protein crystals form.
Reference:
- Li L, Du W, and Ismagilov RF. Multiparameter screening on SlipChip used for nanoliter protein crystallization combining free interface diffusion and microbatch methods. J Am Chem Soc. 2010 Jan 13;132(1):112-9. PMID: 20000709.



