 Science Highlights
Science Highlights
Accelerating a Paradigm Shift
For the Public
The behavior and function of individual cells in the body can vary greatly, even  between cells that are very close together. These differences can play a role in determining health, disease, and therapy outcomes, making the ability to study single cells crucial. The NIH Common Fund Single Cell Analysis Program (SCAP) launched in 2012 to speed up the discovery, development, and translation of approaches to analyze single cells in humans. A variety of new tools and technologies for single cell analysis were developed through the program. SCAP funding ended in 2017. During the period of SCAP funding, there was a significant increase in interest and funding for “single cell analysis” studies, indicating that the program accelerated research in the field. The technological advances made possible by SCAP will undoubtedly have a broad impact on health and disease research.
between cells that are very close together. These differences can play a role in determining health, disease, and therapy outcomes, making the ability to study single cells crucial. The NIH Common Fund Single Cell Analysis Program (SCAP) launched in 2012 to speed up the discovery, development, and translation of approaches to analyze single cells in humans. A variety of new tools and technologies for single cell analysis were developed through the program. SCAP funding ended in 2017. During the period of SCAP funding, there was a significant increase in interest and funding for “single cell analysis” studies, indicating that the program accelerated research in the field. The technological advances made possible by SCAP will undoubtedly have a broad impact on health and disease research.
For Researchers
Variation in cells in human tissues can play a role in determining health, disease, and therapeutic outcomes, making the ability to analyze single cells critical. The NIH Common Fund Single Cell Analysis Program (SCAP) launched in 2012 to speed up the discovery, development, and translation of approaches to analyze single cells in humans. SCAP particularly focused on technology development and a variety of single cell technologies were developed to analyze DNA sequence, DNA methylation, chromosome conformation, and chromatin state. Technology development evolved around three broad themes: droplet-based sequencing approaches, enhanced spatial resolution via fluorescent-based techniques, and barcoding techniques to multiplex microscopic approaches. In 2014, SCAP instituted a grand challenge, called “Follow that Cell,” to stimulate development of new tools for analyzing changes in individual cells over time. The winning project demonstrated a new nanopipette technology that can be used to repeatedly and non-destructively monitor the molecular properties of single cells over time. In addition, the program established the SCAP Transcriptome Consortium project, which developed a public portal containing phenotypic information and whole transcriptome data from 56 human subjects. SCAP-developed resources have been used by the Genotype-Tissue Expression (GTEx) program and the Human BioMolecular Atlas Program (HuBMAP), among others. SCAP funding ended in 2017. The technological advances made possible by SCAP, particularly in single cell RNA-seq and multiplexed imaging combined with computational methods, will undoubtedly have a broad impact on health and disease research.
Reference:
Accelerating a Paradigm Shift- the Common Fund Single Cell Analysis Program. Roy AL, Conroy R, Smith J, Yao Y, Beckel-Mitchener AC, Anderson JM, and Wilder EL. Science Advances. August 2018.
 New Single Cell System for Studying Live Human Brain Cells
New Single Cell System for Studying Live Human Brain Cells
 A team of scientists, led by Single Cell Analysis Program investigator Dr. James Eberwine, have developed a new system for studying live human brain cells donated from neurosurgery, which will help researchers understand human brain diseases and how to treat them. The human brain is made up of an intricate network of different cell types that interact in a variety of ways, with certain diseases and drugs uniquely targeting different brain cell types and their interactions. Single cell analysis can be used to unlock the mystery of differences between these distinct brain cell (neuronal) populations. Although there are existing methods for studying individual neurons, they cannot be used for long term studies using samples from human brain – an important step towards a personalized approach to understanding and treating brain disorders. This new system developed by Dr. Eberwine’s team, using donated cells from seven patients, allowed researchers to grow adult human neurons in the laboratory for long term studies (up to three weeks). Single cell analysis was then carried out on 300 living neurons. Researchers analyzed RNA from individual neurons, which allowed them to determine which genes were “turned on,” or expressed, in each cell. From this analysis, they were able to show that over 12,000 genes were expressed. Interestingly, they found instances where hundreds of genes were only expressed in certain cell types and the cells obtained from each donor had a different pattern of gene expression. Understanding differences in gene expression could potentially be used to develop personalized approaches for evaluating and treating patients with a variety of brain diseases.
A team of scientists, led by Single Cell Analysis Program investigator Dr. James Eberwine, have developed a new system for studying live human brain cells donated from neurosurgery, which will help researchers understand human brain diseases and how to treat them. The human brain is made up of an intricate network of different cell types that interact in a variety of ways, with certain diseases and drugs uniquely targeting different brain cell types and their interactions. Single cell analysis can be used to unlock the mystery of differences between these distinct brain cell (neuronal) populations. Although there are existing methods for studying individual neurons, they cannot be used for long term studies using samples from human brain – an important step towards a personalized approach to understanding and treating brain disorders. This new system developed by Dr. Eberwine’s team, using donated cells from seven patients, allowed researchers to grow adult human neurons in the laboratory for long term studies (up to three weeks). Single cell analysis was then carried out on 300 living neurons. Researchers analyzed RNA from individual neurons, which allowed them to determine which genes were “turned on,” or expressed, in each cell. From this analysis, they were able to show that over 12,000 genes were expressed. Interestingly, they found instances where hundreds of genes were only expressed in certain cell types and the cells obtained from each donor had a different pattern of gene expression. Understanding differences in gene expression could potentially be used to develop personalized approaches for evaluating and treating patients with a variety of brain diseases.
In the News:
- First cell culture of live adult human neurons shows potential of brain cell types, Adult Human Neurons Grown in the Laboratory for the First Time
Reference:
- Primary Cell Culture of Live Neurosurgically Resected Aged Adult Human Brain Cells and Single Cell Transcriptomics. Spaethling JM, Na YJ, Lee J, Ulyanova AV, Baltuch GH, Bell TJ, Brem S, Chen HI, Dueck H, Fisher SA, Garcia MP, Khaladkar M, Kung DK, Lucas TH Jr, O'Rourke DM, Stefanik D, Wang J, Wolf JA, Bartfai T, Grady MS, Sul JY, Kim J, Eberwine JH. Cell Reports. January 2017.
 Spatial Complexity of Mouse Hippocampus Resolved Using seqFISH
Spatial Complexity of Mouse Hippocampus Resolved Using seqFISH
 Single cell analysis has shown that all cells of a particular “type” are not identical – individual cells within the same population can differ dramatically. Understanding these differences will uncover fundamental biological principles and ultimately improve the detection and treatment of diseases. New approaches are needed that study the spatial organization of tissues at single cell resolution and overcome limitations in current measurement methods. Toward this aim, Single Cell Analysis grantee Dr. Long Cai, Assistant Professor of Chemistry at the California Institute of Technology, identified unique gene expression states in the mouse hippocampus, a part of the brain that plays an important role in memory, by quantifying and clustering 249 genes in 16,958 cells. This was accomplished using sequential barcoded fluorescence in situ hybridization (seqFISH), a technique that correlates the spatial expression of different genes in the same cell without removing cells from their original location within tissue. Using seqFISH, the authors report resolving the structural organization of the mouse hippocampus with unprecedented single cell resolution. Further, they found that although some regions are relatively homogenous at a single cell level, others have a high degree of cellular heterogeneity. This work is important because it shows the potential for seqFISH to become a widely used tool to address key concepts in biomedical research, including expanding our knowledge of the molecular anatomy of the brain.
Single cell analysis has shown that all cells of a particular “type” are not identical – individual cells within the same population can differ dramatically. Understanding these differences will uncover fundamental biological principles and ultimately improve the detection and treatment of diseases. New approaches are needed that study the spatial organization of tissues at single cell resolution and overcome limitations in current measurement methods. Toward this aim, Single Cell Analysis grantee Dr. Long Cai, Assistant Professor of Chemistry at the California Institute of Technology, identified unique gene expression states in the mouse hippocampus, a part of the brain that plays an important role in memory, by quantifying and clustering 249 genes in 16,958 cells. This was accomplished using sequential barcoded fluorescence in situ hybridization (seqFISH), a technique that correlates the spatial expression of different genes in the same cell without removing cells from their original location within tissue. Using seqFISH, the authors report resolving the structural organization of the mouse hippocampus with unprecedented single cell resolution. Further, they found that although some regions are relatively homogenous at a single cell level, others have a high degree of cellular heterogeneity. This work is important because it shows the potential for seqFISH to become a widely used tool to address key concepts in biomedical research, including expanding our knowledge of the molecular anatomy of the brain.
Reference:
- In situ transcription profiling of single cells reveals spatial organization of cells in the mouse hippocampus. Shah S, Lubeck E, Zhou W, Cai L. Neuron. October 2016.
 Resolving Dense Gene Expression Profiling Using corrFISH
Resolving Dense Gene Expression Profiling Using corrFISH
 Messenger RNAs (mRNAs) are key biological intermediates in transcribing and translating the genetic code and therefore can be used to measure gene expression. Single cell biology to follow gene expression in individual cells, in their native environment, is important for the advancement of several fields of biomedical research. Single molecule fluorescence in situ hybridization (smFISH) is a recently developed method that uses labeled fluorescent probes to target specific mRNAs. This results in cells that have fluorescent spots, due to hybridization of probes, where each spot corresponds to a single mRNA. These spots can then be used to measure levels of gene expression in single cells. Sequential barcoding fluorescence in situ hybridization (seqFISH) uses smFISH to label multiple mRNAs, using sequential rounds of FISH probes to “barcode” mRNAs. Although this method allows for the detection of a large number of mRNAs directly in cells or tissues at a single cell level, challenges can arise in resolving the barcodes due to the density of the spots and high background. To address this challenge, Single Cell Analysis researcher Dr. Long Cai, Assistant Professor at the California Institute of Technology, developed the correlation FISH (corrFISH) method. This method uses the correlation of images resulting from different seqFISH barcode conditions to resolve high density mRNA barcodes. With corrFISH, the Cai group was able to quantify gene expression and spatial location in single cultured cells and mouse thymus sections. The authors conclude that “in principle, this image correlation approach can be applied to decode high-density images to multiplex molecular species other than RNA, including proteins and metabolites”.
Messenger RNAs (mRNAs) are key biological intermediates in transcribing and translating the genetic code and therefore can be used to measure gene expression. Single cell biology to follow gene expression in individual cells, in their native environment, is important for the advancement of several fields of biomedical research. Single molecule fluorescence in situ hybridization (smFISH) is a recently developed method that uses labeled fluorescent probes to target specific mRNAs. This results in cells that have fluorescent spots, due to hybridization of probes, where each spot corresponds to a single mRNA. These spots can then be used to measure levels of gene expression in single cells. Sequential barcoding fluorescence in situ hybridization (seqFISH) uses smFISH to label multiple mRNAs, using sequential rounds of FISH probes to “barcode” mRNAs. Although this method allows for the detection of a large number of mRNAs directly in cells or tissues at a single cell level, challenges can arise in resolving the barcodes due to the density of the spots and high background. To address this challenge, Single Cell Analysis researcher Dr. Long Cai, Assistant Professor at the California Institute of Technology, developed the correlation FISH (corrFISH) method. This method uses the correlation of images resulting from different seqFISH barcode conditions to resolve high density mRNA barcodes. With corrFISH, the Cai group was able to quantify gene expression and spatial location in single cultured cells and mouse thymus sections. The authors conclude that “in principle, this image correlation approach can be applied to decode high-density images to multiplex molecular species other than RNA, including proteins and metabolites”.
Reference:
- Dense transcript profiling in single cells by image correlation decoding. Coskun A, Cai L. Nature Methods. June 2016.
 Single Cell Analysis Reveals Neuronal Subtypes of the Human Brain
Single Cell Analysis Reveals Neuronal Subtypes of the Human Brain
 The human brain contains around a hundred billion neurons densely packed into three pounds of tissue. Neurons differ from one another in structure, function, and genetic landscape. Although this complex neuronal diversity is fundamental to human brain function, it is still not known how many different types of neurons there are in the brain. This lack of knowledge has impeded our understanding of how the brain functions in health and human disease. Toward understanding brain neuron diversity, Single Cell Analysis Program researcher Dr. Kun Zhang and collaborators examined the gene expression profiles of individual neurons, isolated from post mortem human brain tissue, using single cell RNA sequencing. 3227 sets of single-neuron data from six distinct regions of the cerebral cortex were generated and 16 neuronal subtypes were identified. The authors report that this robust and scalable method lays the “groundwork for high-throughput global human brain transcriptome mapping”. This approach could provide insight into brain function, and potentially disease mechanisms, through distinct profiles of gene expression signatures.
The human brain contains around a hundred billion neurons densely packed into three pounds of tissue. Neurons differ from one another in structure, function, and genetic landscape. Although this complex neuronal diversity is fundamental to human brain function, it is still not known how many different types of neurons there are in the brain. This lack of knowledge has impeded our understanding of how the brain functions in health and human disease. Toward understanding brain neuron diversity, Single Cell Analysis Program researcher Dr. Kun Zhang and collaborators examined the gene expression profiles of individual neurons, isolated from post mortem human brain tissue, using single cell RNA sequencing. 3227 sets of single-neuron data from six distinct regions of the cerebral cortex were generated and 16 neuronal subtypes were identified. The authors report that this robust and scalable method lays the “groundwork for high-throughput global human brain transcriptome mapping”. This approach could provide insight into brain function, and potentially disease mechanisms, through distinct profiles of gene expression signatures.
Reference:
- Neuronal subtypes and diversity revealed by single-nucleus RNA sequencing of the human brain. Lake BB. et al. Science. June 2016.
Read additional commentary in The Scientist: Single-Cell RNA Sequencing Reveals Neuronal Diversity.
 A Tale of Two Alleles
A Tale of Two Alleles
 Genomic imprinting is an epigenetic phenomenon in which one copy of an inherited gene is imprinted to be turned “off”. Under some circumstances, such as embryonic development and various disease states, the imprinted gene can be turned “on” and is expressed. This is known as biallelic expression, when both maternal and paternal alleles of an imprinted gene are expressed. Although imprinting defects are important in health and human disease, the contribution of allele-specific expression in single cells within a population is unclear. For example, it is not known whether imprinted gene expression patterns in complex tissue are cell-type specific or whether individual cells express either one or both alleles of a given gene. Toward answering these questions, Single Cell Analysis researcher Dr. Arjun Raj worked with collaborator Dr. Marisa S. Bartolomei to measure allele-specific expression in single cells, using single-nucleotide polymorphism RNA fluorescent in situ hybridization (SNP FISH). Interestingly, mouse models showed allele-specific expression at the single cell level, in both embryonic fibroblast cells and in cardiac tissue. Their results show that two distinct subpopulations exist: one bilallelically expressing both alleles and one expressing only a maternal copy. This work is important because it shows that defects in genomic imprinting can occur via profound cell-to-cell differences, providing a potential explanation for the pathology associated with human imprinting disorders.
Genomic imprinting is an epigenetic phenomenon in which one copy of an inherited gene is imprinted to be turned “off”. Under some circumstances, such as embryonic development and various disease states, the imprinted gene can be turned “on” and is expressed. This is known as biallelic expression, when both maternal and paternal alleles of an imprinted gene are expressed. Although imprinting defects are important in health and human disease, the contribution of allele-specific expression in single cells within a population is unclear. For example, it is not known whether imprinted gene expression patterns in complex tissue are cell-type specific or whether individual cells express either one or both alleles of a given gene. Toward answering these questions, Single Cell Analysis researcher Dr. Arjun Raj worked with collaborator Dr. Marisa S. Bartolomei to measure allele-specific expression in single cells, using single-nucleotide polymorphism RNA fluorescent in situ hybridization (SNP FISH). Interestingly, mouse models showed allele-specific expression at the single cell level, in both embryonic fibroblast cells and in cardiac tissue. Their results show that two distinct subpopulations exist: one bilallelically expressing both alleles and one expressing only a maternal copy. This work is important because it shows that defects in genomic imprinting can occur via profound cell-to-cell differences, providing a potential explanation for the pathology associated with human imprinting disorders.
Reference:
- Visualizing allele-specific expression in single cells reveals epigenetic mosaicism in an H19 loss-of-imprinting mutant. Ginart P, Kalish JM, Jiang CL, Yu AC, Bartolomei MS, Raj A. Genes and Development. March 2016.
 Keeping Genomic Elements in the Picture
Keeping Genomic Elements in the Picture
 The genomic information of individual cells in the human body is not exactly the same and can change over time, as observed in aging, cancer, and viral infection. These changes can alter the functional organization of the human genome and interactions between genomic elements, resulting in altered gene expression. Yet, the detection of genomic interactions in single living cells remains a major challenge for research scientists. To address this challenge, Single Cell researcher Dr. Bo Huang and collaborators have further developed CRISPR-Cas9 imaging technology to label genomic elements for microscopy detection. This approach allows for the imaging of multiple genomic elements in the same single cell, with potential applications in monitoring events related to human disease.
The genomic information of individual cells in the human body is not exactly the same and can change over time, as observed in aging, cancer, and viral infection. These changes can alter the functional organization of the human genome and interactions between genomic elements, resulting in altered gene expression. Yet, the detection of genomic interactions in single living cells remains a major challenge for research scientists. To address this challenge, Single Cell researcher Dr. Bo Huang and collaborators have further developed CRISPR-Cas9 imaging technology to label genomic elements for microscopy detection. This approach allows for the imaging of multiple genomic elements in the same single cell, with potential applications in monitoring events related to human disease.
Reference:
- Expanding the CRIPSR imaging toolset with Staphylococcus aureus Cas9 for simultaneous imaging of multiple genomic loci. Chen B, Hu J, Almeida R, Liu H, Balakrishnan S, Covill-Cooke C, Lim WA, Huang, B. Nucleic Acids Res. January 2016.
 Single-Cell Analysis: Powerful Drops in the Buckets
Single-Cell Analysis: Powerful Drops in the Buckets
 Single Cell researcher Marc Kirschner has developed new single cell analysis technology called inDrop. inDrop is capable of analyzing very small tissue samples while capturing a greater percentage of cells than other technology. Kirschner and colleagues used inDrop to analyze thousands of differentiated and embryonic stem cells from mice. More news: Harvard Groups Tap Microfluidics for Single-Cell RNA-Seq Methods.
Single Cell researcher Marc Kirschner has developed new single cell analysis technology called inDrop. inDrop is capable of analyzing very small tissue samples while capturing a greater percentage of cells than other technology. Kirschner and colleagues used inDrop to analyze thousands of differentiated and embryonic stem cells from mice. More news: Harvard Groups Tap Microfluidics for Single-Cell RNA-Seq Methods.
Reference:
- Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Klein AM, Mazutis L, Akartuna I, Tallapragada N, Veres A, Li V, Peshkin L, Weitz DA, Kirschner MW. Cell. May 2015.
 Understanding Tumor Genomic Diversity Using Single Cell Analysis
Understanding Tumor Genomic Diversity Using Single Cell Analysis
 Cancer is caused by small changes or mutations to DNA within cells. Although next-generation DNA sequencing can identify common mutations in a population of cancer cells, there are limited resources available for studying the genomes of individual cells that make up a tumor. This is important, given that individual cells from the same tumor can show distinct mutational characteristics, which may present challenges when designing effective cancer treatment strategies. To gain insight into the genomic diversity of breast cancer tumors, Dr. Navin Nicholas’ research team at the University of Texas MD Anderson Cancer Center has developed a single-cell whole genome sequencing technique, called Nuc-Seq. Using this method, Dr. Navin’s group reports that no two individual tumor cells are genetically identical. This technique will allow for a better understanding of genetic diversity within individual tumors and has the potential to influence our approach to cancer treatment.
Cancer is caused by small changes or mutations to DNA within cells. Although next-generation DNA sequencing can identify common mutations in a population of cancer cells, there are limited resources available for studying the genomes of individual cells that make up a tumor. This is important, given that individual cells from the same tumor can show distinct mutational characteristics, which may present challenges when designing effective cancer treatment strategies. To gain insight into the genomic diversity of breast cancer tumors, Dr. Navin Nicholas’ research team at the University of Texas MD Anderson Cancer Center has developed a single-cell whole genome sequencing technique, called Nuc-Seq. Using this method, Dr. Navin’s group reports that no two individual tumor cells are genetically identical. This technique will allow for a better understanding of genetic diversity within individual tumors and has the potential to influence our approach to cancer treatment.
Reference:
- Clonal evolution in breast cancer revealed by single nucleus genome sequencing. Wang Y, Waters J, Leung ML, Unruh A, Roh W, Shi X, Chen K, Scheet P, Vattathil S, Liang H, Multani A, Zhang H, Zhao R, Michor F, Meric-Bernstam F, Navin NE. Nature. August 2014.
 Single Cell Analysis Researchers Create 3D Movies of Neuronal Activity in Living Animals
Single Cell Analysis Researchers Create 3D Movies of Neuronal Activity in Living Animals
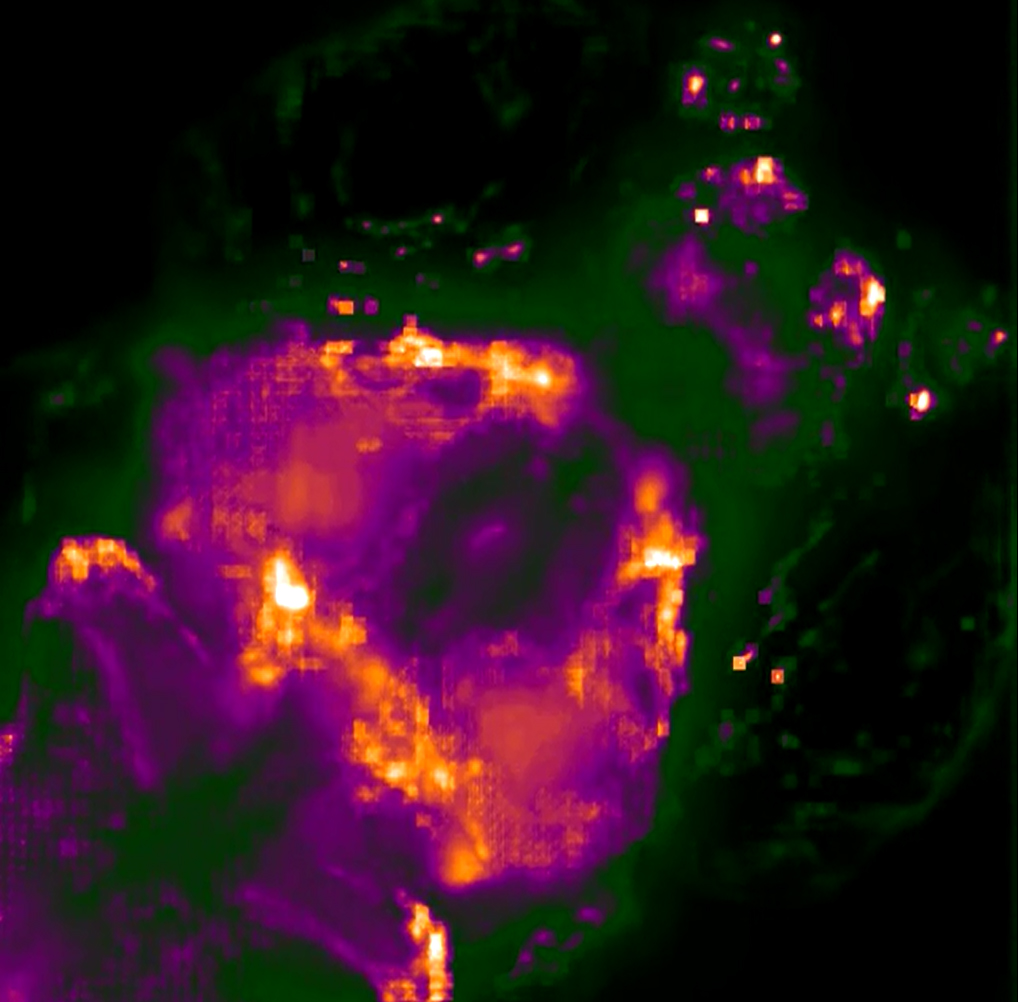 Researchers in the Common Fund’s Single Cell Analysis program have created a high-speed, large-scale 3D imaging system capable of visualizing the activity of individual neurons in a living animal. The technique created by Dr. Edward Boyden and colleagues at the Massachusetts Institute of Technology (MIT) and the University of Vienna can generate three-dimensional (3D) movies of entire brains at the millisecond timescale, the first time this has ever been accomplished. The new method optimizes a widely used technology known as light-field microscopy (LFM), which creates 3D images by measuring the angles of incoming rays of light. Boyden and colleagues sent the light emitted by the sample being imaged through an array of lenses, which refracted the light in different directions and generated about 400 different points of light. Using a computer algorithm, the points of light were recombined to generate the 3D structure of the sample. Using this new system they were able to simultaneously image the activity of every neuron in the worm Caenorhabditis elegans and the whole brain of a zebrafish larva expressing proteins that fluoresce when binding to calcium (a way to visualize neuron activation). The technique offers a more complete picture of nervous system activity than has previously been possible. Through monitoring and tracking neurons and neural pathways, scientists hope to discover how sensory input is processed and behavior generated.
Researchers in the Common Fund’s Single Cell Analysis program have created a high-speed, large-scale 3D imaging system capable of visualizing the activity of individual neurons in a living animal. The technique created by Dr. Edward Boyden and colleagues at the Massachusetts Institute of Technology (MIT) and the University of Vienna can generate three-dimensional (3D) movies of entire brains at the millisecond timescale, the first time this has ever been accomplished. The new method optimizes a widely used technology known as light-field microscopy (LFM), which creates 3D images by measuring the angles of incoming rays of light. Boyden and colleagues sent the light emitted by the sample being imaged through an array of lenses, which refracted the light in different directions and generated about 400 different points of light. Using a computer algorithm, the points of light were recombined to generate the 3D structure of the sample. Using this new system they were able to simultaneously image the activity of every neuron in the worm Caenorhabditis elegans and the whole brain of a zebrafish larva expressing proteins that fluoresce when binding to calcium (a way to visualize neuron activation). The technique offers a more complete picture of nervous system activity than has previously been possible. Through monitoring and tracking neurons and neural pathways, scientists hope to discover how sensory input is processed and behavior generated.
Reference:
- Simultaneous Whole-Animal 3D Imaging of Neuronal Activity Using Light-Field Microscopy. Prevedel R, Yoon Y-G, Hoffmann M, Pak N, Wetzstein G, Kato S, Schrödel T, Raskar R, Zimmer M, Boyden ES, and Vaziri A. Nature Methods. July 2014.
 Single-cell Phenotyping Within Transparent Intact Tissue Through Whole-Body Clearing
Single-cell Phenotyping Within Transparent Intact Tissue Through Whole-Body Clearing
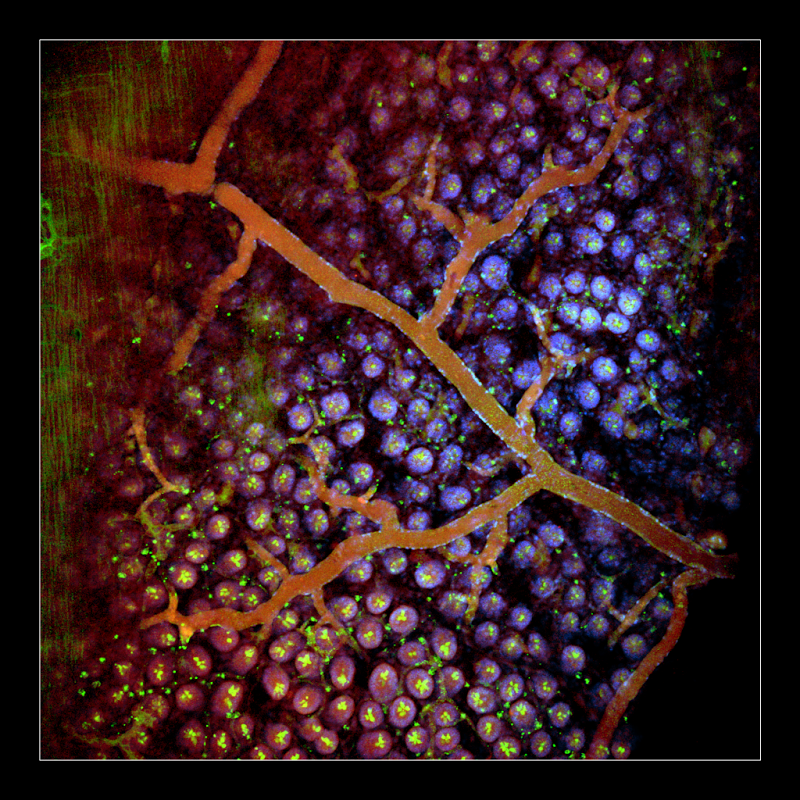 The unavailability of 3D structural maps for many organs makes understanding the structure-function relationships at cellular, circuit and organ-wide scale difficult. This knowledge gap has been attributed to the absence of a method that enables whole-organ imaging. In this publication, several techniques are described that allow for the process of tissue clearing during which whole organs and bodies are rendered optically transparent, thereby exposing their cellular structure with intact connectivity. The techniques that allow for this whole-organ imaging include PACT (passive clarity technique) which allows for passive tissue clearing and staining of intact organs; RIMS (refractive index matching solution) which is a mounting media for imaging thick tissue; and PARS (perfusion-assisted agent release in situ) which is a method for whole-body clearing and labeling. This group of scientists was able to show in rodents that these techniques are compatible with the endogenous fluorescence, other more well-established techniques, subcellular resolution and long-term storage. These methods are applicable for high-resolution and content mapping of both normal and abnormal elements within intact organs and bodies, paving the way to the understanding structure-function relationships on a whole organ or animal scale.
The unavailability of 3D structural maps for many organs makes understanding the structure-function relationships at cellular, circuit and organ-wide scale difficult. This knowledge gap has been attributed to the absence of a method that enables whole-organ imaging. In this publication, several techniques are described that allow for the process of tissue clearing during which whole organs and bodies are rendered optically transparent, thereby exposing their cellular structure with intact connectivity. The techniques that allow for this whole-organ imaging include PACT (passive clarity technique) which allows for passive tissue clearing and staining of intact organs; RIMS (refractive index matching solution) which is a mounting media for imaging thick tissue; and PARS (perfusion-assisted agent release in situ) which is a method for whole-body clearing and labeling. This group of scientists was able to show in rodents that these techniques are compatible with the endogenous fluorescence, other more well-established techniques, subcellular resolution and long-term storage. These methods are applicable for high-resolution and content mapping of both normal and abnormal elements within intact organs and bodies, paving the way to the understanding structure-function relationships on a whole organ or animal scale.
Reference:
- Single-Cell Phenotyping within Transparent Intact Tissue through Whole-Body Clearing. Yang B, Treweek JB, Kulkarni RP, Deverman BE, Chen CK, Lubeck E, Shah S, Cai L, Gradinaru V. Cell. August 2014.
Examining Genetic Function and Regulation at the Systems Level using TIVA
 Multicellular organisms are comprised of various different types of cells that are categorized based on their function and phenotypic expression. These cells, however, may not be identical at the molecular level and can be more heterogeneous in terms of their mRNA composition and proteins. Most of the knowledge about variability in genes has come from studies involving unicellular organisms, but it is unknown whether the processes that control variability in gene expression in these single-celled organisms can translate to the cells of the multicellular organisms. The environment in which cells exist can be diverse and have an effect on the gene expression. It is therefore of great biological importance to explore the RNA profiles of cells while the cell is in its natural environment. RNA sequencing allows for the exploration of a single cell’s pool of mRNA, however, the profiling of the set of RNA molecules within a cell (transcriptome) while the cell is within its microenvironment is dependent upon methods that are noninvasive and precise. Dr. Eberwine and others at the Penn Genome Frontiers Institute have developed a tag which upon photoactivation enables the mRNA capture from single cells in live tissue. Using this transcriptome in vivo analysis (TIVA) tag combined with traditional RNA sequencing, they have been able to analyze the variance in transcriptome among single neurons in culture as well as mouse and human tissue in its natural state (uninterrupted).
Multicellular organisms are comprised of various different types of cells that are categorized based on their function and phenotypic expression. These cells, however, may not be identical at the molecular level and can be more heterogeneous in terms of their mRNA composition and proteins. Most of the knowledge about variability in genes has come from studies involving unicellular organisms, but it is unknown whether the processes that control variability in gene expression in these single-celled organisms can translate to the cells of the multicellular organisms. The environment in which cells exist can be diverse and have an effect on the gene expression. It is therefore of great biological importance to explore the RNA profiles of cells while the cell is in its natural environment. RNA sequencing allows for the exploration of a single cell’s pool of mRNA, however, the profiling of the set of RNA molecules within a cell (transcriptome) while the cell is within its microenvironment is dependent upon methods that are noninvasive and precise. Dr. Eberwine and others at the Penn Genome Frontiers Institute have developed a tag which upon photoactivation enables the mRNA capture from single cells in live tissue. Using this transcriptome in vivo analysis (TIVA) tag combined with traditional RNA sequencing, they have been able to analyze the variance in transcriptome among single neurons in culture as well as mouse and human tissue in its natural state (uninterrupted).
Reference:
- Transcriptome in vivo analysis (TIVA) of spatially defined single cells in live tissue. Lovatt D, Ruble BK, Lee J, Dueck H, Kim TK, Fisher S, Francis C, Spaethling JM, Wolf JA, Grady MS, Ulyanova AV, Yeldell SB, Griepenburg JC, Buckley PT, Kim J, Sul JY, Dmochowski IJ, Eberwine J. Nature Methods. February 2014.
 Sequencing RNA Where it Lives by FISSEQ
Sequencing RNA Where it Lives by FISSEQ
 A group of scientists at Harvard Medical School previously described a method of sequencing, using fluorescence, without disturbing neighboring cells (in situ), termed FISSEQ, in 2003. This technique allows for the sequencing of libraries fixed in gel or on a glass slide and produces 8bp of sequence. This method, just like other methods, are limited to a handful of genes per specimen, making it costly and time-intensive to localize all the RNA molecules (transcriptome) within any given cell, let alone an entire specimen. In a 2014 publication, they describe a new and improved version of FISSEQ that is capable of sequencing the RNA transcriptome. In this study, they examined the RNA expression and localization in human cells (fibroblasts) that synthesize the extracellular matrix and collagen of tissues and determined it to be compatible the examination of tissues sections and even an entire embryo. They were also able to demonstrate imaging and analytic capabilities of this technique across multiple types of specimens. In the future, they expect to further improve this technique by increasing read length of sequences, coverage, and library preparation, which might lead to more refined identification of diseased tissues in clinical medicine.
A group of scientists at Harvard Medical School previously described a method of sequencing, using fluorescence, without disturbing neighboring cells (in situ), termed FISSEQ, in 2003. This technique allows for the sequencing of libraries fixed in gel or on a glass slide and produces 8bp of sequence. This method, just like other methods, are limited to a handful of genes per specimen, making it costly and time-intensive to localize all the RNA molecules (transcriptome) within any given cell, let alone an entire specimen. In a 2014 publication, they describe a new and improved version of FISSEQ that is capable of sequencing the RNA transcriptome. In this study, they examined the RNA expression and localization in human cells (fibroblasts) that synthesize the extracellular matrix and collagen of tissues and determined it to be compatible the examination of tissues sections and even an entire embryo. They were also able to demonstrate imaging and analytic capabilities of this technique across multiple types of specimens. In the future, they expect to further improve this technique by increasing read length of sequences, coverage, and library preparation, which might lead to more refined identification of diseased tissues in clinical medicine.
Reference:
- Highly multiplexed subcellular RNA sequencing in situ.Lee JH, Daugharthy ER, Scheiman J, Kalhor R, Yang JL, Ferrante TC, Terry R, Jeanty SS, Li C, Amamoto R, Peters DT, Turczyk BM, Marblestone AH, Inverso SA, Bernard A, Mali P, Rios X, Aach J, Church GM.Science. March 2014.
Massively Parallel Polymerase Cloning and Genome Sequencing of Single Cells Using Nanoliter Microwells
 Genome sequencing of single cells has a variety of applications, including characterizing difficult-to-culture microorganisms and identifying somatic mutations in single cells from mammalian tissues. A major hurdle in this process is the bias in amplifying the genetic material from a single cell, a procedure known as polymerase cloning. Here we describe the microwell displacement amplification system (MIDAS), a massively parallel polymerase cloning method in which single cells are randomly distributed into hundreds to thousands of nanoliter wells and their genetic material is simultaneously amplified for shotgun sequencing.
Genome sequencing of single cells has a variety of applications, including characterizing difficult-to-culture microorganisms and identifying somatic mutations in single cells from mammalian tissues. A major hurdle in this process is the bias in amplifying the genetic material from a single cell, a procedure known as polymerase cloning. Here we describe the microwell displacement amplification system (MIDAS), a massively parallel polymerase cloning method in which single cells are randomly distributed into hundreds to thousands of nanoliter wells and their genetic material is simultaneously amplified for shotgun sequencing.
Image courtesy of Nature Publishing Group
Cellular Barcodes for Single Cell Profiling
 Dr. Christopher Love at the Massachusetts Institute of Technology and colleagues have developed techniques to label heterogeneous or homogeneous cells using “barcoding” (i.e. combinatorial application of dyes, streptavidin/biotin or antibody labels), sort the barcoded cells into small wells, and then characterize the cytokines (signaling molecules) produced by the cell(s) in each of the wells. The characterization of cytokines is accomplished by a novel method, termed “microengraving,” previously developed by the authors and which uses a panel of cytokine-specific antibodies attached to a glass slide placed over the array of wells to detect cytokines. Barcoding can dramatically increase throughput of analysis and decrease reagent requirements. The authors demonstrate the utility of this approach in several different applications including constructing dose-response curves, profiling secretory responses to a variety of stimuli, and profiling secretory responses as a function of cell lineage. An intriguing, but yet unrealized, application of this method is the quantitative analysis of the communication among members in a small, well-defined group of cells contained in a single well. Such analysis may reveal fundamental insights into cell-cell interactions not obtainable through traditional methods.
Dr. Christopher Love at the Massachusetts Institute of Technology and colleagues have developed techniques to label heterogeneous or homogeneous cells using “barcoding” (i.e. combinatorial application of dyes, streptavidin/biotin or antibody labels), sort the barcoded cells into small wells, and then characterize the cytokines (signaling molecules) produced by the cell(s) in each of the wells. The characterization of cytokines is accomplished by a novel method, termed “microengraving,” previously developed by the authors and which uses a panel of cytokine-specific antibodies attached to a glass slide placed over the array of wells to detect cytokines. Barcoding can dramatically increase throughput of analysis and decrease reagent requirements. The authors demonstrate the utility of this approach in several different applications including constructing dose-response curves, profiling secretory responses to a variety of stimuli, and profiling secretory responses as a function of cell lineage. An intriguing, but yet unrealized, application of this method is the quantitative analysis of the communication among members in a small, well-defined group of cells contained in a single well. Such analysis may reveal fundamental insights into cell-cell interactions not obtainable through traditional methods.
Reference:
- Yamanaka YJ, Szeto GL, Gierahn TM, Forcier TL, Benedict KF, Brefo MS, Lauffenburger DA, Irvine DJ, Love JC. Cellular barcodes for efficiently profiling single-cell secretory responses by microengraving. Anal Chem. 2012 Dec 18;84(24):10531-6. PMID: 23205933



