Science Highlights
Signatures for Success: Reducing Side Effects of Cancer Drugs
 The right drug can help stop cancer in its tracks by preventing cell growth and blocking the formation of new blood vessels—cutting off the supply of nutrients that the cancer needs in order to survive and grow. However, sometimes cancer drugs can also cause unwanted side effects, like damage to the heart (cardiotoxicity), which can impair blood flow throughout the body. The Library of Integrated Network-based Cellular Signatures (LINCS) Drug Toxicity Signature Generation Center (DToxS) used a technique called transcriptomic profiling combined with clinical data and drug structure data to develop a new way to predict if cancer drugs might cause cardiotoxicity in patients.
The right drug can help stop cancer in its tracks by preventing cell growth and blocking the formation of new blood vessels—cutting off the supply of nutrients that the cancer needs in order to survive and grow. However, sometimes cancer drugs can also cause unwanted side effects, like damage to the heart (cardiotoxicity), which can impair blood flow throughout the body. The Library of Integrated Network-based Cellular Signatures (LINCS) Drug Toxicity Signature Generation Center (DToxS) used a technique called transcriptomic profiling combined with clinical data and drug structure data to develop a new way to predict if cancer drugs might cause cardiotoxicity in patients.
LINCS researchers used human heart cells (cardiomyocytes) to test 23 different FDA-approved cancer drugs, collecting thousands of data points using the transcriptomic profiling technique to show how each different drug changed the types and amounts of molecules produced by the heart cells. This analysis allowed the scientists to pinpoint the specific combinations of molecular changes that might be related to cardiotoxicity. In parallel, they examined the structures of the cancer drugs and analyzed clinical data to see if the molecular changes found in the heart cells could also be found in cancer patients treated with these medications. Taking all this together, they found that testing cancer drugs in human heart cells along with an analysis of the structure of the drug could be combined with clinical data to create a drug ‘signature’ to help predict unwanted side effects like cardiotoxicity. This drug signature approach developed by the LINCS research team could be a useful way to assess safety early in the drug development process, potentially identifying the medications that have a lower chance of causing unwanted side effects.
The protocols and data sets generated by this research are available in the DToxS Data Repository and are freely available for research use.
Reference: Transcriptomic profiling of human cardiac cells predicts protein kinase inhibitor-associated cardiotoxicity. van Hasselt JGC, Rahman R, Hansen J, Stern A, Shim JV, Xiong Y, Pickard A, Jayaraman G, Hu B, Mahajan M, Gallo JM, Goldfarb J, Sobie EA, Birtwistle MR, Schlessinger A, Azeloglu EU, Iyengar R. Nat Commun. 2020 Sep 23;11(1):4809.
Signature Improvement: Decoding Breast Cancer Drug Targets
![]() A study from the NIH Common Fund’s Library of Integrated Network-based Cellular Signatures (LINCS) program has uncovered why some metastatic breast cancer drugs may work better than others. Metastatic breast cancer, also called ‘Stage 4’ breast cancer, occurs when the cancer has spread to other parts of the body such as the lungs, liver, bones, or brain. A team at LINCS awardee institution Harvard Medical School, led by Dr. Peter Sorger, and their collaborators analyzed three drugs (palbociclib, robociclib, and abemaciclib) that work by blocking a cancer cell’s ability to grow and multiply. All three drugs were thought to function in a similar way, but it was unclear whether each drug may also have unique differences in how they block cancer cells that might affect which drug should be chosen to treat different patients.
A study from the NIH Common Fund’s Library of Integrated Network-based Cellular Signatures (LINCS) program has uncovered why some metastatic breast cancer drugs may work better than others. Metastatic breast cancer, also called ‘Stage 4’ breast cancer, occurs when the cancer has spread to other parts of the body such as the lungs, liver, bones, or brain. A team at LINCS awardee institution Harvard Medical School, led by Dr. Peter Sorger, and their collaborators analyzed three drugs (palbociclib, robociclib, and abemaciclib) that work by blocking a cancer cell’s ability to grow and multiply. All three drugs were thought to function in a similar way, but it was unclear whether each drug may also have unique differences in how they block cancer cells that might affect which drug should be chosen to treat different patients.
The researchers used LINCS tools (including Growth Rate Inhibition metrics, CMap, and Enrichr) and analyses at the RNA, protein, and cellular levels to generate a composite ‘drug signature’ for how each of the drugs affected the cancer cells—like a snapshot of how the cells responded to treatment. Interestingly, they found that one of the drugs (abemaciclib) had a broader range of molecular targets that may make it a good candidate for treating breast cancers that have become resistant to therapy. 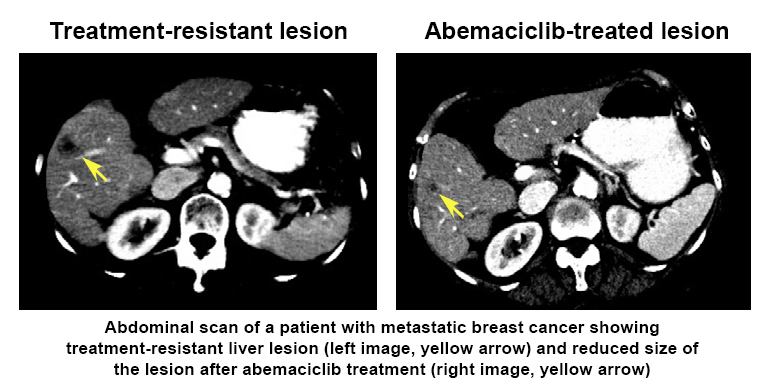 Further, they showed that abemaciclib treatment of one patient’s drug-resistant metastatic breast cancer decreased the size of a cancer lesion in the patient’s liver. This research highlights how drug signatures may help tailor treatment to improve patient outcomes and identifies targets for developing effective and longer lasting treatments for drug-resistant metastatic breast cancers.
Further, they showed that abemaciclib treatment of one patient’s drug-resistant metastatic breast cancer decreased the size of a cancer lesion in the patient’s liver. This research highlights how drug signatures may help tailor treatment to improve patient outcomes and identifies targets for developing effective and longer lasting treatments for drug-resistant metastatic breast cancers.
The data sets generated by this research are available in the HMS LINCS Database and are freely available for research use.
Reference: Multiomics Profiling Establishes the Polypharmacology of FDA-Approved CDK4/6 Inhibitors and the Potential for Differential Clinical Activity. Hafner M, Mills CE, Subramanian K, Chen C, Chung M, Boswell SA, Everley RA, Liu C, Walmsley CS, Juric D, Sorger PK. Cell Chem Biol. 2019 Aug 15; 26(8):1067-1080.
LINCS in Action: Reducing Side Effects of Anti-Inflammatory Drugs Used to Treat Skin Conditions
The Library of Integrated Network-based Cellular Signatures (LINCS) contains a wealth of information about how cells respond to a variety of drugs. One of the ways researchers are using this information is to reverse engineer desired cell responses. The first step is to identify how a cell’s responses are altered in a specific disease or condition, then LINCS can be used to find a drug that may help counteract that response and switch the cell back to a healthy state. This process is also being used to optimize treatment by reducing or preventing harmful side effects of drugs. Using this strategy, researchers can identify promising drug candidates, and then test them in cells or model systems.
Glucocorticoids are anti-inflammatory drugs that are routinely used to treat a variety of diseases and conditions, including eczema and psoriasis. Unfortunately, continuous treatment with glucocorticoids can harm the treated tissues. When these drugs are used topically to treat skin conditions it can lead to thinning of the skin, and may also cause increased skin fragility, bruising, skin infections, and delayed wound healing. Researchers turned to LINCS to help identify drugs that could counteract the harmful side effects caused by long term use of glucocorticoids, while preserving the beneficial effects of the drug.
An earlier study found that treating skin with glucocorticoids caused a specific cell response—increased activity of two genes, REDD1 and FKBP51. 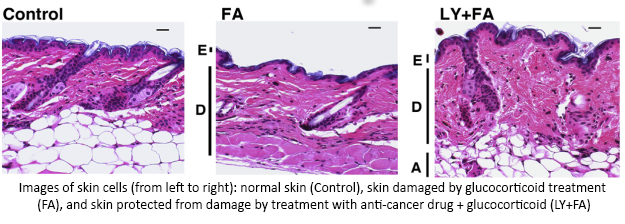 Using LINCS, researchers discovered a class of cancer-fighting drugs that could help make glucocorticoid treatments safer by reducing activity of REDD1 and FKBP51. Using a mouse model, they showed that combining glucocorticoids with one of these anti-cancer drugs protected skin from damage and reduced thinning. This combination allowed for the positive effects of the treatment while minimizing the harmful side effects. Additional testing is needed to determine if this approach could work in humans and could help reduce other unwanted side effects, such as delayed wound healing and increased risk of skin infections. This use of LINCS illustrates the power of LINCS information to improve the design of tailored treatments—reducing the harmful side effects of drugs without losing the benefits.
Using LINCS, researchers discovered a class of cancer-fighting drugs that could help make glucocorticoid treatments safer by reducing activity of REDD1 and FKBP51. Using a mouse model, they showed that combining glucocorticoids with one of these anti-cancer drugs protected skin from damage and reduced thinning. This combination allowed for the positive effects of the treatment while minimizing the harmful side effects. Additional testing is needed to determine if this approach could work in humans and could help reduce other unwanted side effects, such as delayed wound healing and increased risk of skin infections. This use of LINCS illustrates the power of LINCS information to improve the design of tailored treatments—reducing the harmful side effects of drugs without losing the benefits.
This work was not financially supported by LINCS.
Reference: PI3K inhibitors protect against glucocorticoid-induced skin atrophy. Agarwal S, Mirzoeva S, Readhead B, Dudley JT, Budunova I. EBioMedicine. 2019 Mar; 41:526-537.
LINCS In Action: Discovering a New Treatment Approach for Alzheimer's Disease
The Library of Integrated Network-based Cellular Signatures program (LINCS) is developing a library of molecular signatures that
 describes how different types of cells respond to a variety of agents that disrupt normal cellular functions. Currently LINCS provides nearly 400 data sets and over fifty freely available online tools useful for a broad range of research applications, including studies to understand disease processes and predict health outcomes. These LINCS resources are also being used in basic research studies to help develop new therapies for a variety of diseases, including Alzheimer’s disease.
describes how different types of cells respond to a variety of agents that disrupt normal cellular functions. Currently LINCS provides nearly 400 data sets and over fifty freely available online tools useful for a broad range of research applications, including studies to understand disease processes and predict health outcomes. These LINCS resources are also being used in basic research studies to help develop new therapies for a variety of diseases, including Alzheimer’s disease.
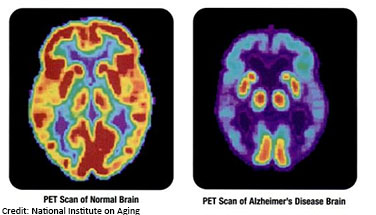
A hallmark of Alzheimer’s disease is chronic neuroinflammation (inflammation of the brain) that, over time, damages the neurons critical for normal brain function. Neuroinflammation also has an important role in maintaining health -- acting like a cleaning service for removing infections and molecular debris from the brain. An ideal treatment for Alzheimer’s disease would protect neurons from damage while still allowing for the cellular cleanup provided by neuroinflammation. The LINCS KINOMEscan resource is being used to help scientists develop new approaches to treat neurological disorders like Alzheimer’s disease while still preserving the functional benefits of neuroinflammation.
KINOMEscan is a database of drug interactions that helps scientists predict what targets drugs will bind to, and the strength of that interaction. Research funded by NINDS and NIA coupled a cell-based screening assay with KINOMEscan data, which led to the discovery of a new drug that works by inhibiting two different proteins involved in damaging neuroinflammation – GSK3β and CDK5. Using a zebrafish model of Alzheimer’s disease, the scientists demonstrated that the drug directly protects neurons from neuroinflammation-induced damage without preventing the health-promoting effects of neuroinflammation. This promising discovery may also have implications for other neurodegenerative diseases involving neuroinflammation, such as amyotrophic lateral sclerosis (ALS), and illustrates the power of LINCS data and resources for driving exciting new research findings to help improve health.
This work was not financially supported by LINCS.
Reference:Dual Inhibition of GSK3β and CDK5 Protects the Cytoskeleton of Neurons from Neuroinflammatory-Mediated Degeneration In Vitro and In Vivo. Reinhardt L, Kordes S, Reinhardt P, Glatza M, Baumann M, Drexler HCA, Menninger S, Zischinsky G, Eickhoff J, Fröb C, Bhattarai P, Arulmozhivarman G, Marrone L, Janosch A, Adachi K, Stehling M, Anderson EN, Abo-Rady M, Bickle M, Pandey UB, Reimer MM, Kizil C, Schöler HR, Nussbaumer P, Klebl B, Sterneckert JL. Stem Cell Reports. 2019 Mar 5;12(3):502-517.
BD2K-LINCS Data Science Symposium (DSS 2018)
The third annual BD2K-LINCS Data Science Symposium was held at the University of Miami on January 31-February 2, 2018. The symposium focused on the study of complex biological systems using large-scale cellular perturbation profiling and applications in drug development, translational biomedicine, and environmental health. Check out the archived videos of the presentation on the BD2K-LINCS DCIC YouTube Channel.
The Connectivity Map: An Online Public Resource for Gene Activity Profiles
 One common method for profiling the state of a cell is to measure levels of gene expression, the process by which the information within our genes is used to make a gene product. Cells control expression of genes as a way to adapt to their environment.
One common method for profiling the state of a cell is to measure levels of gene expression, the process by which the information within our genes is used to make a gene product. Cells control expression of genes as a way to adapt to their environment.
Researchers at the LINCS Transcriptomics Center have released an updated version of the Connectivity Map (CMap) which now contains over 1.3 million gene expression profiles from treating numerous types of human cells with over 42,000 agents that disrupt cellular function (perturbagens). This extremely large set of data is based on the L1000 assay which allows for inexpensive and large-scale analysis of gene expression.
CMap is a freely available public resource that any research scientist can use and already has more than 18,000 registered users. For example, researchers can gain insight regarding how a small-molecule drug works in a cell by determining the changes in gene expression it produces. Alternatively, they can find a molecule from all those that have been screened that has a desired effect in cells to aid in the design of new therapeutics.
One example of the utility of the L1000 assay is a collaboration within the LINCS Consortium in which gene expression data were combined with a method for cell growth and survival measurements developed at the Harvard Medical School LINCS Center. These measures were used to profile the effects of 109 small-molecule drugs on six different breast cancer cell lines. This study showed there is substantial variability in how certain drugs affect the cell growth and survival certain types of breast cancer cells. These differences can be explored to help us understand why some patients respond better to breast cancer treatments than others. LINCS researchers also demonstrated that gene expression results could be used to predict combinations of drugs that are effective in preventing growth of the breast cancer cells, which could potentially be used to design combination therapies.
References:
A Next Generation Connectivity Map: L1000 Platform and the First 1,000,000 Profiles. Subramanian A, Narayan R, Corsello SM, Peck DD, Natoli TE, Lu X, Gould J, Davis JF, Tubelli AA, Asiedu JK, Lahr DL, Hirschman JE, Liu Z, Donahue M, Julian B, Khan M, Wadden D, Smith IC, Lam D, Liberzon A, Toder C, Bagul M, Orzechowski M, Enache OM, Piccioni F, Johnson SA, Lyons NJ, Berger AH, Shamji AF, Brooks AN, Vrcic A, Flynn C, Rosains J, Takeda DY, Hu R, Davison D, Lamb J, Ardlie K, Hogstrom L, Greenside P, Gray NS, Clemons PA, Silver S, Wu X, Zhao WN, Read-Button W, Wu X, Haggarty SJ, Ronco LV, Boehm JS, Schreiber SL, Doench JG, Bittker JA, Root DE, Wong B, Golub TR. Cell. 2017 Nov 30; 171(6): 1437-1452.
Common and cell-type specific responses to anti-cancer drugs revealed by high throughput transcript profiling. Niepel M, Hafner M, Duan Q, Wang Z, Paull EO, Chung M, Lu X, Stuart JM, Golub TR, Subramanian A, Ma'ayan A, Sorger PK. Nat Commun. 2017 Oct 30; 8(1): 1186.
Using LINCS and other Big Data to Find New Cancer Treatments
A new study using computational tools to combine multiple sets of previously generated biomedical big data has identified drugs that can be repurposed to fight cancer. Dr. Bin Chen, a Big Data to Knowledge (BD2K) Mentored Career Development Awardee, and colleagues took advantage of multiple existing data sets, including the NIH Common Fund’s Library of Integrated Network-based Cellular Signatures (LINCS) L1000 data set and the National Cancer Institute’s The Cancer Genome Atlas (TCGA) data set to generate a list of potential drugs predicted to inhibit the growth of liver cancer cells. Dr. Chen and colleagues compared the global cellular changes between normal and cancerous liver cells in the TCGA data set with changes caused by treating cells with a panel of 12,442 distinct chemical compounds in the LINCS data sets. They tested the hypothesis that compounds causing inverse changes to those seen when a cell becomes cancerous would be strong candidates for treating cancer. Using this approach, four compounds were selected that had the potential to reverse the changes seen in liver cancer patients. When these compounds were tested against liver cancer cell lines grown in the laboratory, all four inhibited the growth of these cells. The most potent of these drugs was also tested in a mouse liver cancer model and was shown to reduce the size of tumors. This drug (pyrvinium pamoate), which has been previously FDA approved to treat pinworm infections, will require further experimentation to determine if it can be repurposed to treat liver cancer in humans.
The growing number of large, robust experimental data sets in biomedical research offers unprecedented opportunities for new discoveries using computational approaches, which is a continuing goal of the BD2K program. This study, also demonstrates the power of using the LINCS data sets to discover novel therapeutics by focusing on global cellular changes instead of a singular molecular target. Similar approaches have the potential to yield candidate drugs for repurposing to treat a wide spectrum of diseases.
Read the press release from UCSF here.
Reference:
Reversal of cancer gene expression correlates with drug efficacy and reveals therapeutic targets. Chen B, Ma L, Paik H1, Sirota M, Wei W, Chua MS, So S, Butte AJ. Nat Commun. 2017 Jul 12. 8:16022.
Announcing the Connectivity Map Challenge
 The Common Fund-supported LINCS Center for Transcriptomics, in partnership with the Crowd Innovation Lab at Harvard Business School, launched its first challenge, “Infer the Transcriptome.” Contestants were provided with a large dataset of ~100,000 gene expression profiles on which to train an inference model. Models are scored based on their accuracy in predicting gene expression values for non-landmark genes in a separate test dataset. The contest format is a 2-week marathon featuring a continuously updated leader board. To determine winners, each contestant’s best model will be scored on its performance on a holdout dataset. The challenge began June 28, 2016.
The Common Fund-supported LINCS Center for Transcriptomics, in partnership with the Crowd Innovation Lab at Harvard Business School, launched its first challenge, “Infer the Transcriptome.” Contestants were provided with a large dataset of ~100,000 gene expression profiles on which to train an inference model. Models are scored based on their accuracy in predicting gene expression values for non-landmark genes in a separate test dataset. The contest format is a 2-week marathon featuring a continuously updated leader board. To determine winners, each contestant’s best model will be scored on its performance on a holdout dataset. The challenge began June 28, 2016.
For More Information:
Connectivity Map Challenge information and leader board
Crowd Innovation Lab at Harvard Business School
Topcoder Platform
Contacts: Jin Paik - [email protected], Aravind Subramanian - [email protected]
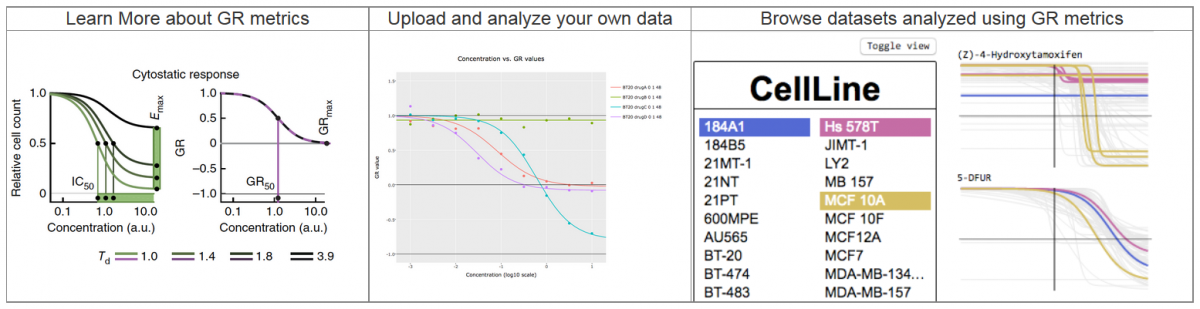
Introducing the Growth Rate Calculator and Browser
Drug-response studies play an important role in both preclinical and clinical research, but such studies are complicated by differences in cell growth rates across samples and conditions. To improve the value and reliability of such studies, new metrics for measuring drug response were developed and published in Nature Methods by the HMS LINCS Center. The online GR tools were developed by the BD2K-LINCS Data Coordination and Integration Center.
Reference:
Growth rate inhibition metrics correct for confounders in measuring sensitivity to cancer drugs. Hafner M, Niepel M, Chung M, Sorger PK. Growth rate inhibition metrics correct for confounders in measuring sensitivity to cancer drugs. Nature Methods 2016.
Large-Scale Analysis of Cellular Signaling
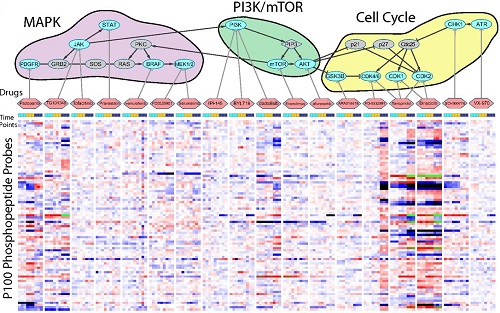 Congratulations to LINCS grantee Dr. Jacob Jaffe! Dr. Jaffe's research linking modified proteins, cell signaling, and drug-induced changes in cellular states was recently featured on the cover of the journal Molecular & Cellular Proteomics. Read the article abstract.
Congratulations to LINCS grantee Dr. Jacob Jaffe! Dr. Jaffe's research linking modified proteins, cell signaling, and drug-induced changes in cellular states was recently featured on the cover of the journal Molecular & Cellular Proteomics. Read the article abstract.
Reference:
Reduced-representation Phosphosignatures Measured by Quantitative Targeted MS Capture Cellular States and Enable Large-scale Comparison of Drug-induced Phenotypes. Abelin JG, Patel J, Lu X, Feeney CM, Fagbami L, Creech AL, Hu R, Lam D, Davison D, Pino L, Qiao JW, Kuhn E, Officer A, Li J, Abbatiello S, Subramanian A, Sidman R, Snyder E, Carr SA, Jaffe JD. Molecular & Cellular Proteomics. 2016 May; 15(5): 1622-41.
LINCS and Big Data in Genetics
![]() Learn how high-throughput data from the LINCS program could contribute to our understanding of cancer and other diseases in this editorial from Nature (note that full text may require institutional journal access).
Learn how high-throughput data from the LINCS program could contribute to our understanding of cancer and other diseases in this editorial from Nature (note that full text may require institutional journal access).
Network Analysis and Systems Biology Massive Open Online Course
 A Network Analysis and Systems Biology Massive Open Online Course was hosted by the BD2K-LINCS Data Coordination and Integration Center on Coursera February 29 - May 16, 2016. Visit the Coursera website to learn more about the course.
A Network Analysis and Systems Biology Massive Open Online Course was hosted by the BD2K-LINCS Data Coordination and Integration Center on Coursera February 29 - May 16, 2016. Visit the Coursera website to learn more about the course.
LINCS Scientists Investigate Sex Differences
 Sex and gender play a role in how health and disease differ across individuals, and considering these factors during research informs the development of preventive and therapeutic interventions for both sexes. Visit the Sex as a Biological Variable webpage to learn how supplements to LINCS grants enable researchers to investigate sex as a biological variable.
Sex and gender play a role in how health and disease differ across individuals, and considering these factors during research informs the development of preventive and therapeutic interventions for both sexes. Visit the Sex as a Biological Variable webpage to learn how supplements to LINCS grants enable researchers to investigate sex as a biological variable.
LINCS Data Harnessed to Help Reveal How Cancer Cells Continuously Reproduce
 Cancer, generally speaking, occurs due to unregulated cell reproduction. As cells grow and multiply unchecked they need to produce more proteins, ultimately leading to the formation of tumors. While it has long been known that cancer cells increase their production of proteins, the interface between production of proteins and gene expression in cells has remained unclear. In a 2013 study published in the journal Science, a research team used complex analyses to better understand these interactions. Through the use of Common Fund supported LINCS data, the researchers were able to show that HSF1, a critical cell regulator, was highly controlled by the production of proteins. The LINCS data used for these analyses are a large catalog of how human cells react at the genetic level to changes induced by a vast array of chemicals or interfering genetic molecules. The analyses of the data showed that the response of cells to an agent that blocks proteins from being made is similar to their response when a protein that controls genes is blocked, suggesting that this protein (HSF1) regulates the production of proteins in the cells. This is a key finding in better understanding why cancer cells continue to have energy, while ordinary cells would otherwise be worn out. HSF1 has long been known to be a critical regulator in cancer, playing a large role in maintaining uncontrolled growth. Therefore understanding how HSF1 is controlled is an important finding in the search for improved cancer treatments. In the study, the research team was able to use this information to reverse HSF1 activation and suppress tumor growth in a mouse model. In the model, human leukemia cells were grafted into the mice, showing that the effects have a potentially promising role in humans. These results are very encouraging for future studies that could impact a number of tumor-forming cancers affecting humans.
Cancer, generally speaking, occurs due to unregulated cell reproduction. As cells grow and multiply unchecked they need to produce more proteins, ultimately leading to the formation of tumors. While it has long been known that cancer cells increase their production of proteins, the interface between production of proteins and gene expression in cells has remained unclear. In a 2013 study published in the journal Science, a research team used complex analyses to better understand these interactions. Through the use of Common Fund supported LINCS data, the researchers were able to show that HSF1, a critical cell regulator, was highly controlled by the production of proteins. The LINCS data used for these analyses are a large catalog of how human cells react at the genetic level to changes induced by a vast array of chemicals or interfering genetic molecules. The analyses of the data showed that the response of cells to an agent that blocks proteins from being made is similar to their response when a protein that controls genes is blocked, suggesting that this protein (HSF1) regulates the production of proteins in the cells. This is a key finding in better understanding why cancer cells continue to have energy, while ordinary cells would otherwise be worn out. HSF1 has long been known to be a critical regulator in cancer, playing a large role in maintaining uncontrolled growth. Therefore understanding how HSF1 is controlled is an important finding in the search for improved cancer treatments. In the study, the research team was able to use this information to reverse HSF1 activation and suppress tumor growth in a mouse model. In the model, human leukemia cells were grafted into the mice, showing that the effects have a potentially promising role in humans. These results are very encouraging for future studies that could impact a number of tumor-forming cancers affecting humans.
Reference
Santagata S, Mendillo ML, Tang YC, Subramanian A, Perley CC, Roche SP, Wong B, Narayan R, Kwon H, Koeva M, Amon A, Golub TR, Porco JA Jr, Whitesell L, Lindquist S. Tight coordination of protein translation and HSF1 activation supports the anabolic malignant state. Science, July 19, 2013; 341. PMID: 23869022.
LINCS Researchers Find New Insights into Measures of Drug Activity
More effective therapeutics are needed for numerous conditions affecting people, yet drug development remains inefficient. Investigators supported by the Common Fund Library of Integrated Network-based Cellular Signatures (LINCS) program have discovered new insights into the pharmacological properties of drugs that should help us design better therapeutics and more accurately predict their effects. While most studies of drug effects focus on measures of drug potency, e.g., by emphasizing the dose needed to experimentally reduce cell numbers in half, Fallahi-Sichani and colleagues at Harvard Medical School and the Oregon Health and Science University found that other measures of the response of cells to drugs can provide additional insights. They found that different measures are more informative at different doses of the drug response, e.g., some are more informative at higher doses and some at lower doses. They also found that the different measures do not always correlate with each other, e.g., when compared across different drugs or different cell types. Yet some measures correlate with cell type and others with drug class. In addition, the different measures each reveal unique information that contributes insights into the action of the drug. The findings indicate that it is worthwhile to compare multiple parameters when examining the variability of drug effects, and expand the way we should think about parameters of drug activity. In some cases the underlying explanation comes from how individual cells might behave differently from a population of cells.
References:
Fallahi-Sichani M, Honarnejad S, Heiser LM, Gray JW, Sorger PK. Metrics other than potency reveal systematic variation in responses to cancer drugs. Nat Chem Biol. 2013 Sep 8. PMID 24013279.
Common Fund Announces Awards for the LINCS Program
The Common Fund is supporting two awards totaling $12.7 million dollars in the Library of Integrated Network-based Cellular Signatures (LINCS) program. The awards support the high-throughput collection and integrative computational analysis of molecular activity and cellular signatures generated in response to a variety of perturbing agents in a multiple cell types. The new knowledge generated by this program will serve as a long-term resource for the scientific community.
- One new award, given to Drs. Todd Golub and Wendy Winckler of the Broad Institute, supports the application of a novel approach to analysis of genome-wide expression to catalogue the cellular consequences of 4,000 diverse small molecules and genetic perturbations in an array of twenty different human cell types representing biological diversity and interest in the broad scientific community. The work will establish the information resource needed to support the discovery of unknown components of the genome, annotate the function of small molecules, and link disease states with small-molecule or signatures of genetic perturbation to provide insight into the biological basis of disease and potential new therapeutics (1U54-HG006093-01)
- The second award given to Drs. Timothy Mitchinson and Peter Sorger of Harvard University, supports the development of a new research center to advance the knowledge of disease processes, drug mechanisms and selectivity, and ultimately patient-specific responses to therapy. The center will focus on small molecule kinase inhibitors as versatile perturbagens which may be targeted in new therapies. The researchers will characterize the response to these perturbagens in 45 different cell lines known for diverse drug responses and for which genomic data are available, and then in more than 1000 human tumor cell lines (1U54-HG006097-01)
Overcoming Obstacles to Analyzing Complex Biological Data
 Biomedical research data generated from genomics analyses, imaging, biochemistry and other assays are abundant yet difficult to integrate using conventional approaches and databases. To address this need, Dr. Peter Sorger and colleagues at Harvard Medical School, Massachusetts Institute of Technology, and the University of Applied Sciences in Germany, researchers supported through the Common Fund’s Library of Integrated Network-based Cellular Signatures (LINCS) program, have developed an innovative new adaptable method that allows different types of complex data sets to be stored, analyzed and extended. In a recent paper in Nature Methods, they demonstrate the utility of the approach, which exploits useful aspects of two data file formats (HDF5 and XML), for analyzing a complex imaging data set reflecting 160 experimental conditions in over a million different single cells. The approach led to the discovery of new pharmacological relationships between compounds that bind and inhibit epidermal growth factor receptors (EGFR), providing insights into cell-to-cell variability in response to drugs.
Biomedical research data generated from genomics analyses, imaging, biochemistry and other assays are abundant yet difficult to integrate using conventional approaches and databases. To address this need, Dr. Peter Sorger and colleagues at Harvard Medical School, Massachusetts Institute of Technology, and the University of Applied Sciences in Germany, researchers supported through the Common Fund’s Library of Integrated Network-based Cellular Signatures (LINCS) program, have developed an innovative new adaptable method that allows different types of complex data sets to be stored, analyzed and extended. In a recent paper in Nature Methods, they demonstrate the utility of the approach, which exploits useful aspects of two data file formats (HDF5 and XML), for analyzing a complex imaging data set reflecting 160 experimental conditions in over a million different single cells. The approach led to the discovery of new pharmacological relationships between compounds that bind and inhibit epidermal growth factor receptors (EGFR), providing insights into cell-to-cell variability in response to drugs.
References:
Millard BL, Niepel M, Menden MP, Muhlich JL, and Sorger PK. Adaptive informatics for multifactorial and high-content biological data. Nat Methods. 2011. Vol 8(6):487-493.